1DDE
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, YTTRIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DD9
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, STRONTIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
2FL7
 
 | | S. cerevisiae Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Keck, J.L, Hou, Z, Daner, J.R, Fox, C.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Protein Sci., 15, 2006
|
|
1TXY
 
 | | E. coli PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Keck, J.L, Lopper, M, Holton, J.M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PriB, a component of the Escherichia coli replication restart primosome
Structure, 12, 2004
|
|
3PLW
 
 | | Ref protein from P1 bacteriophage | | Descriptor: | Recombination enhancement function protein, SULFATE ION, ZINC ION | | Authors: | Keck, J.L, Lu, D, Cox, M.M. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Creating Directed Double-strand Breaks with the Ref Protein: A NOVEL RecA-DEPENDENT NUCLEASE FROM BACTERIOPHAGE P1.
J.Biol.Chem., 286, 2011
|
|
2RHF
 
 | | D. radiodurans RecQ HRDC domain 3 | | Descriptor: | DNA helicase RecQ, PHOSPHATE ION | | Authors: | Keck, J.L, Killoran, M.P. | | Deposit date: | 2007-10-09 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ.
Nucleic Acids Res., 36, 2008
|
|
3C95
 
 | | Exonuclease I (apo) | | Descriptor: | Exodeoxyribonuclease I, MAGNESIUM ION | | Authors: | Keck, J.L, Lu, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3C9B
 
 | | Crystal structure of SeMet Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Keck, J.L, Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
4TMU
 
 | |
7R7J
 
 | | Crystal structure of RadD with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | Authors: | Osorio Garcia, M.A, Satyshur, K.A, Keck, J.L, Cox, M.M. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the Escherichia coli RadD DNA repair protein bound to ADP reveals a novel zinc ribbon domain.
Plos One, 17, 2022
|
|
5JRJ
 
 | | Crystal Structure of Herbaspirillum seropedicae RecA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Leite, W.C, Galvao, C.W, Saab, S.C, Iulek, J, Etto, R.M, Steffens, M.B.R, Chitteni-Pattu, S, Stanage, T, Keck, J.L, Cox, M.M. | | Deposit date: | 2016-05-06 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of H. seropedicae RecA Protein - Insights into the Polymerization of RecA Protein as Nucleoprotein Filament.
PLoS ONE, 11, 2016
|
|
1D3Y
 
 | | STRUCTURE OF THE DNA TOPOISOMERASE VI A SUBUNIT | | Descriptor: | DNA TOPOISOMERASE VI A SUBUNIT, MAGNESIUM ION, SODIUM ION | | Authors: | Nichols, M.D, DeAngelis, K.A, Keck, J.L, Berger, J.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11.
EMBO J., 18, 1999
|
|
1F21
 
 | |
2NCJ
 
 | | Solution Structure of the PriC DNA replication restart protein | | Descriptor: | Uncharacterized protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Wessel, S.R, Keck, J.L, Markley, J.L. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of the PriC DNA Replication Restart Protein.
J.Biol.Chem., 291, 2016
|
|
2OXJ
 
 | | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers: GCN4-p1 with beta-residues at b and f heptad positions. | | Descriptor: | ACETATE ION, hybrid alpha/beta peptide based on the GCN4-p1 sequence; heptad positions b and f substituted with beta-amino acids | | Authors: | Horne, W.S, Price, J.L, Keck, J.L, Gellman, S.H. | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers.
J.Am.Chem.Soc., 129, 2007
|
|
2OXK
 
 | | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers: GCN4-pLI with beta-residues at b and f heptad positions. | | Descriptor: | FORMIC ACID, hybrid alpha/beta peptide based on the GCN4-pLI sequence; heptad positions b and f substituted with beta-amino acids | | Authors: | Horne, W.S, Price, J.L, Keck, J.L, Gellman, S.H. | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers.
J.Am.Chem.Soc., 129, 2007
|
|
4Z0U
 
 | | RNase HI/SSB-Ct complex | | Descriptor: | Ribonuclease H, SSB-Ct Peptide | | Authors: | Petzold, C, Keck, J.L. | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity.
J.Biol.Chem., 290, 2015
|
|
8FAK
 
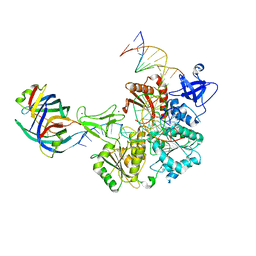 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
3IGM
 
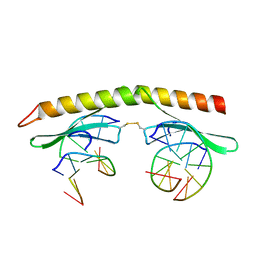 | | A 2.2A crystal structure of the AP2 domain of PF14_0633 from P. falciparum, bound as a domain-swapped dimer to its cognate DNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*TP*GP*CP*A)-3', PF14_0633 protein | | Authors: | Lindner, S.E, De Silva, E, Keck, J.L, Llinas, M. | | Deposit date: | 2009-07-28 | | Release date: | 2009-11-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants of DNA Binding by a P. falciparum ApiAP2 Transcriptional Regulator.
J.Mol.Biol., 395, 2010
|
|
3SXU
 
 | |
3UFM
 
 | |
3UDG
 
 | | Structure of Deinococcus radiodurans SSB bound to ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein, THYMIDINE-5'-PHOSPHATE | | Authors: | George, N.P, Ngo, K.V, Chitteni-Patu, S, Norais, C.A, Battista, J.R, Cox, M.M, Keck, J.L. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes.
J.Biol.Chem., 287, 2012
|
|
3UF7
 
 | |
1WUD
 
 | | E. coli RecQ HRDC domain | | Descriptor: | ATP-dependent DNA helicase recQ | | Authors: | Bernstein, D.A, Keck, J.L. | | Deposit date: | 2004-12-04 | | Release date: | 2005-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conferring Substrate Specificity to DNA Helicases: Role of the RecQ HRDC Domain
STRUCTURE, 13, 2005
|
|
3VDY
 
 | | B. subtilis SsbB/ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein ssbB | | Authors: | Yadav, T, Carrasco, B, Myers, A.R, George, N.P, Keck, J.L, Alonso, J.C. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins.
Nucleic Acids Res., 40, 2012
|
|
