1OGV
 
 | | Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Katona, G, Andreasson, U, Landau, E.M, Andreasson, L.-E, Neutze, R. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lipidic Cubic Phase Crystal Structure of the Photosynthetic Reaction Centre from Rhodobacter Sphaeroides at 2.35 A Resolution
J.Mol.Biol., 331, 2003
|
|
1GVK
 
 | | Porcine pancreatic elastase acyl enzyme at 0.95 A resolution | | Descriptor: | CALCIUM ION, ELASTASE 1, PEPTIDE INHIBITOR, ... | | Authors: | Katona, G, Wilmouth, R.C, Wright, P.A, Berglund, G.I, Hajdu, J, Neutze, R, Schofield, C.J. | | Deposit date: | 2002-02-14 | | Release date: | 2002-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | X-Ray Structure of a Serine Protease Acyl-Enzyme Complex at 0.95-A Resolution.
J.Biol.Chem., 277, 2002
|
|
1H4W
 
 | | Structure of human trypsin IV (brain trypsin) | | Descriptor: | BENZAMIDINE, CALCIUM ION, TRYPSIN IVA | | Authors: | Katona, G, Berglund, G.I, Hajdu, J, Graf, L, Szilagyi, L. | | Deposit date: | 2001-05-15 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure reveals basis for the inhibitor resistance of human brain trypsin.
J. Mol. Biol., 315, 2002
|
|
2BNP
 
 | | Lipidic cubic phase grown reaction centre from Rhodobacter sphaeroides, ground state | | Descriptor: | 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Katona, G, Snijder, A, Gourdon, P, Andreasson, U, Hansson, O, Andreasson, L.E, Neutze, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Regulation of Charge Recombination Reactions in a Photosynthetic Bacterial Reaction Centre
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BNS
 
 | | Lipidic cubic phase grown reaction centre from Rhodobacter sphaeroides, excited state | | Descriptor: | 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Katona, G, Snijder, A, Gourdon, P, Andreasson, U, Hansson, O, Andreasson, L.E, Neutze, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Regulation of Charge Recombination Reactions in a Photosynthetic Bacterial Reaction Centre
Nat.Struct.Mol.Biol., 12, 2005
|
|
2JI3
 
 | | X-ray structure of the iron-peroxide intermediate of superoxide reductase (E114A mutant) from Desulfoarculus baarsii | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (III) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
2JI2
 
 | | X-ray structure of E114A mutant of superoxide reductase from Desulfoarculus baarsii in the native, reduced form | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (II) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
2JI1
 
 | | X-ray structure of wild-type superoxide reductase from Desulfoarculus baarsii | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (II) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
3CLR
 
 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
3CLU
 
 | | Crystal structure of the R236K mutant from Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
3CLT
 
 | | Crystal structure of the R236E mutant of Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
3CLS
 
 | | Crystal structure of the R236C mutant of ETF from Methylophilus methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
8S03
 
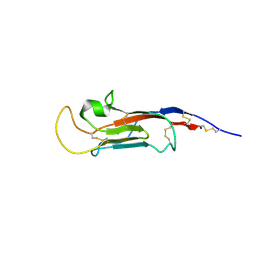 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Method: | SOLUTION NMR | | Cite: | The second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking
To Be Published
|
|
3P8M
 
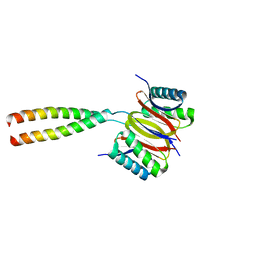 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide dimerized by leucine zipper | | Descriptor: | Dynein light chain 2, General control protein GCN4 | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-10-14 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome.
Plos One, 6, 2011
|
|
6H4E
 
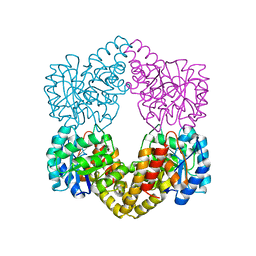 | | Proteus mirabilis N-acetylneuraminate lyase | | Descriptor: | Putative N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Garcia-Bonete, M.J, Goyal, P, Katona, G, Dobson, R.C.J, Friemann, R. | | Deposit date: | 2018-07-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | The structure of Proteus mirabilis N-acetylneuraminate lyase reveals an intermolecular disulphide bond
To Be Published
|
|
4ETA
 
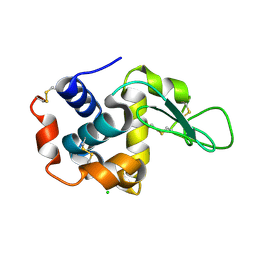 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET8
 
 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETB
 
 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETE
 
 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
2X5V
 
 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
2X5U
 
 | | 80 microsecond Laue diffraction snapshot from crystals of a photosynthetic reaction centre without illumination. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-induced structural changes in a photosynthetic reaction center caught by Laue diffraction.
Science, 328, 2010
|
|
2XTT
 
 | | Bovine trypsin in complex with evolutionary enhanced Schistocerca gregaria protease inhibitor 1 (SGPI-1-P02) | | Descriptor: | ACETATE ION, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Wahlgren, W.Y, Pal, G, Kardos, J, Porrogi, P, Szenthe, B, Patthy, A, Graf, L, Katona, G. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The catalytic aspartate is protonated in the Michaelis complex formed between trypsin and an in vitro evolved substrate-like inhibitor: a refined mechanism of serine protease action.
J.Biol.Chem., 286, 2011
|
|
2XQQ
 
 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide (Ac-SRGTQTE). | | Descriptor: | ACETATE ION, DYNEIN LIGHT CHAIN 2, CYTOPLASMIC, ... | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-09-07 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Directed Evolution Reveals the Binding Motif Preference of the Lc8/Dynll Hub Protein and Predicts Large Numbers of Novel Binders in the Human Proteome
Plos One, 6, 2011
|
|
2F91
 
 | | 1.2A resolution structure of a crayfish trypsin complexed with a peptide inhibitor, SGTI | | Descriptor: | CADMIUM ION, CHLORIDE ION, Serine protease inhibitor I/II, ... | | Authors: | Fodor, K, Harmat, V, Hetenyi, C, Kardos, J, Antal, J, Perczel, A, Patthy, A, Katona, G, Graf, L. | | Deposit date: | 2005-12-05 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme:Substrate Hydrogen Bond Shortening during the Acylation Phase of Serine Protease Catalysis.
Biochemistry, 45, 2006
|
|
5APD
 
 | | Hen Egg White Lysozyme not illuminated with 0.4THz radiation | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
