5H47
 
 | | Crystal structure of AOL complexed with 2-MeSe-Fuc | | Descriptor: | Uncharacterized protein, methyl 6-deoxy-2-Se-methyl-2-seleno-alpha-L-galactopyranoside | | Authors: | Kato, R, Nishikawa, Y, Makyio, H. | | Deposit date: | 2016-10-31 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of seleno-fucose compounds and their application to the X-ray structural determination of carbohydrate-lectin complexes using single/multi-wavelength anomalous dispersion phasing
Bioorg. Med. Chem., 25, 2017
|
|
5EO7
 
 | | Crystal structure of AOL | | Descriptor: | Predicted protein, SULFATE ION, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5EO8
 
 | | Crystal structure of AOL(868) | | Descriptor: | Predicted protein, methyl 1-seleno-beta-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
4OWF
 
 | | Crystal structure of the NEMO CoZi in complex with HOIP NZF1 domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, NF-kappa-B essential modulator, ZINC ION | | Authors: | Rahighi, S, Fujita, H, Kawasaki, M, Kato, R, Iwai, K, Wakatsuki, S. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism Underlying I kappa B Kinase Activation Mediated by the Linear Ubiquitin Chain Assembly Complex.
Mol.Cell.Biol., 34, 2014
|
|
1D2M
 
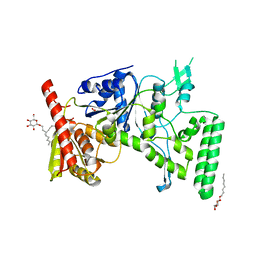 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
3H3S
 
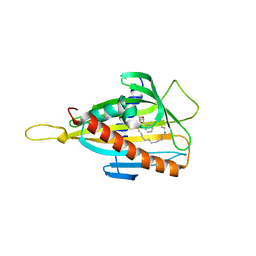 | | Crystal structure of the CERT START domain in complex with HPA-15 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]pentadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3T
 
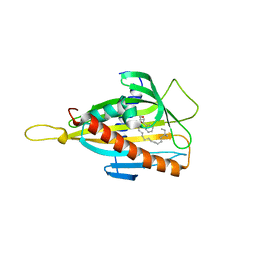 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3Q
 
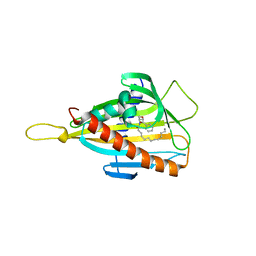 | | Crystal structure of the CERT START domain in complex with HPA-13 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tridecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3R
 
 | | Crystal structure of the CERT START domain in complex with HPA-14 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tetradecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
4DCN
 
 | | Crystal Structure Analysis of the Arfaptin2 BAR domain in Complex with ARL1 | | Descriptor: | ADP-ribosylation factor-like protein 1, Arfaptin-2, MAGNESIUM ION, ... | | Authors: | Nakamura, K, Xie, Y, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for membrane binding specificity of the Bin/Amphiphysin/Rvs (BAR) domain of Arfaptin-2 determined by Arl1 GTPase
J.Biol.Chem., 287, 2012
|
|
1O3Y
 
 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
1O3X
 
 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
1SNT
 
 | | Structure of the human cytosolic sialidase Neu2 | | Descriptor: | Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
4X86
 
 | | Crystal structure of BAG6-Ubl4a complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Large proline-rich protein BAG6, SULFATE ION, ... | | Authors: | Kuwabara, N, Kato, R. | | Deposit date: | 2014-12-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a BAG6 (Bcl-2-associated Athanogene 6)-Ubl4a (Ubiquitin-like Protein 4a) Complex Reveals a Novel Binding Interface That Functions in Tail-anchored Protein Biogenesis
J.Biol.Chem., 290, 2015
|
|
5XFC
 
 | | Serial femtosecond X-ray structure of a stem domain of human O-mannose beta-1,2-N-acetylglucosaminyltransferase solved by Se-SAD using XFEL (refined against 13,000 patterns) | | Descriptor: | 4-nitrophenyl beta-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Fumiaki, Y, Kato, R, Manya, H. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
5XFD
 
 | |
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1WRD
 
 | | Crystal structure of Tom1 GAT domain in complex with ubiquitin | | Descriptor: | Target of Myb protein 1, Ubiquitin | | Authors: | Akutsu, M, Kawasaki, M, Katoh, Y, Shiba, T, Yamaguchi, Y, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain.
Febs Lett., 579, 2005
|
|
5GGF
 
 | |
5GGG
 
 | |
5GGN
 
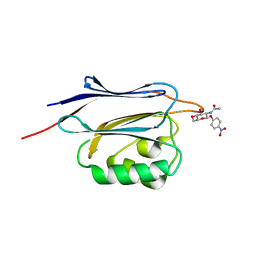 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGJ
 
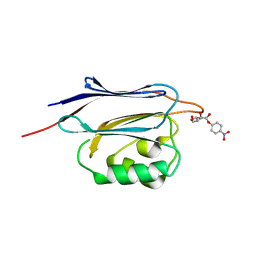 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-alpha-pNP | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGL
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-alpha-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGP
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta1,2-Man-peptide | | Descriptor: | 10-mer Peptide from Dystroglycan, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
