1XHX
 
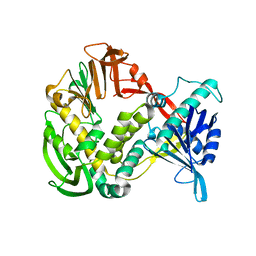 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1XHZ
 
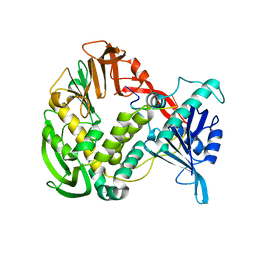 | | Phi29 DNA polymerase, orthorhombic crystal form, ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', DNA polymerase | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
2EX3
 
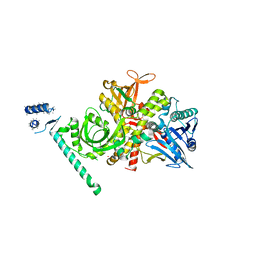 | | Bacteriophage phi29 DNA polymerase bound to terminal protein | | Descriptor: | DNA polymerase, DNA terminal protein, LEAD (II) ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Embo J., 25, 2006
|
|
2GM4
 
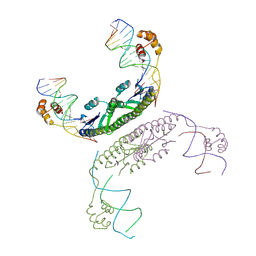 | | An activated, tetrameric gamma-delta resolvase: Hin chimaera bound to cleaved DNA | | Descriptor: | 5'-D(*CP*AP*GP*TP*GP*TP*CP*CP*GP*AP*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*A)-3', 5'-D(*TP*TP*AP*TP*CP*GP*GP*AP*CP*AP*CP*TP*G)-3', Transposon gamma-delta resolvase | | Authors: | Kamtekar, S, Ho, R.S, Li, W, Steitz, T.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GM5
 
 | | An activated, truncated gamma-delta resolvase tetramer | | Descriptor: | Transposon gamma-delta resolvase | | Authors: | Kamtekar, S, Ho, R.S, Li, W, Steitz, T.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NJ2
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus | | Descriptor: | MAGNESIUM ION, Proline-tRNA Synthetase, ZINC ION | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ5
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to proline sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ1
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to cysteine sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ8
 
 | | Crystal Structure of Prolyl-tRNA Synthetase from Methanocaldococcus janaschii | | Descriptor: | Proline-tRNA Synthetase | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ6
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to alanine sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2XR9
 
 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
1XI1
 
 | | Phi29 DNA polymerase ssDNA complex, monoclinic crystal form | | Descriptor: | 5'-D(P*TP*TP*TP*TP*T)-3', DNA polymerase, MAGNESIUM ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of X-ray intensities from single crystals containing lattice-translocation defects
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3LCD
 
 | | Inhibitor Bound to A DFG-In structure of the Kinase Domain of CSF-1R | | Descriptor: | Macrophage colony-stimulating factor 1 receptor, N~3~-(2,6-dichlorobenzyl)-5-(4-{[(2R)-2-(pyrrolidin-1-ylmethyl)pyrrolidin-1-yl]carbonyl}phenyl)pyrazine-2,3-diamine, SULFATE ION | | Authors: | Kamtekar, S, Day, J.E, Reitz, B.A, Mathis, K.J, Meyers, M.J. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based drug design enables conversion of a DFG-in binding CSF-1R kinase inhibitor to a DFG-out binding mode
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LCO
 
 | | Inhibitor Bound to A DFG-Out structure of the Kinase Domain of CSF-1R | | Descriptor: | 3-({4-methoxy-5-[(4-methoxybenzyl)oxy]pyridin-2-yl}methoxy)-5-(1-methyl-1H-pyrazol-4-yl)pyrazin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Kamtekar, S, Day, J.E, Reitz, B.A, Mathis, K.J, Meyers, M.J. | | Deposit date: | 2010-01-11 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based drug design enables conversion of a DFG-in binding CSF-1R kinase inhibitor to a DFG-out binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2ODR
 
 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3KJF
 
 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJN
 
 | | Caspase 8 bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S)-2-{2-[(1H-benzimidazol-5-ylcarbonyl)amino]ethyl}-7-(cyclohexylmethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJQ
 
 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
1HQY
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT1
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
3UJ3
 
 | |
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
2WAP
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with the drug-like urea inhibitor PF-3845 | | Descriptor: | 4-(3-{[5-(trifluoromethyl)pyridin-2-yl]oxy}benzyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Kamtekar, S, Stevens, R.C. | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Highly Selective Faah Inhibitor that Reduces Inflammatory Pain.
Chem.Biol., 16, 2009
|
|
