1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
8V2T
 
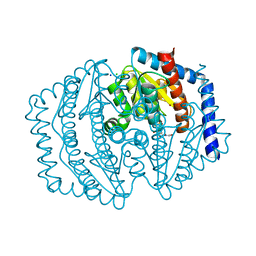 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148591 | | Descriptor: | 1,5,6-trideoxy-6,6-difluoro-1-(N-hydroxyformamido)-6-phosphono-D-ribo-hexitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 2024
|
|
8V4J
 
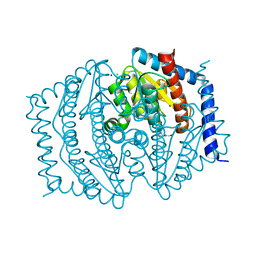 | |
2FQ6
 
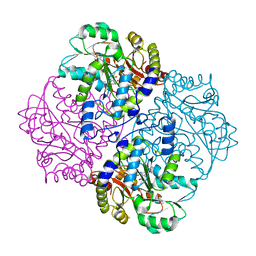 | |
7UDI
 
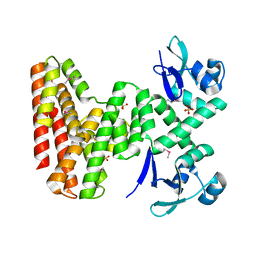 | |
3Q9D
 
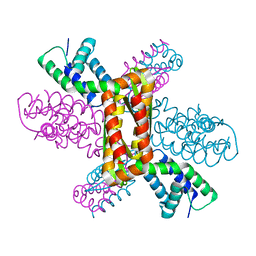 | |
8U1J
 
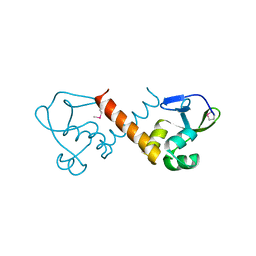 | |
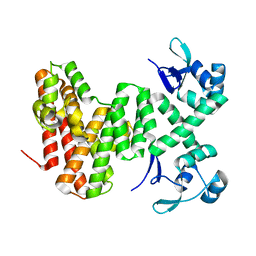
8U0G
 
 | |
6ED7
 
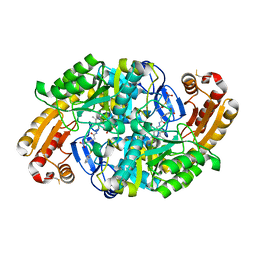 | | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772 | | Descriptor: | 2-[(2-nitrophenyl)sulfanyl]acetohydrazide, 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Brown, C.M, Zlitni, S, Chan, J, Brown, E.D, Junop, M.S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772
To Be Published
|
|
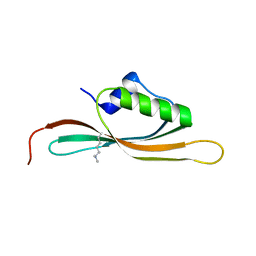
5UVR
 
 | |
3RWR
 
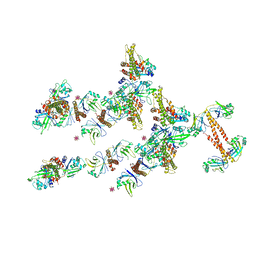 | | Crystal structure of the human XRCC4-XLF complex | | Descriptor: | DNA repair protein XRCC4, HEXATANTALUM DODECABROMIDE, Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2011-05-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.943 Å) | | Cite: | Structure of human XLF-XRCC4: assembly of a functional DNA repair complex
To be Published
|
|
2OPT
 
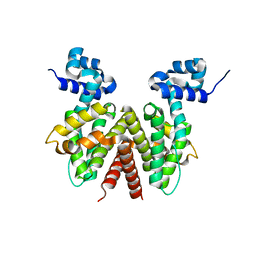 | | Crystal Structure of Apo ActR from Streptomyces coelicolor. | | Descriptor: | ActII protein | | Authors: | Willems, A.R, Junop, M.S. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
2R9A
 
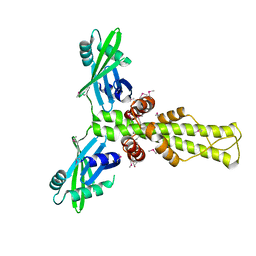 | | Crystal structure of human XLF | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Mol.Cell, 28
|
|
6MC6
 
 | |
6MC8
 
 | |
1ZM0
 
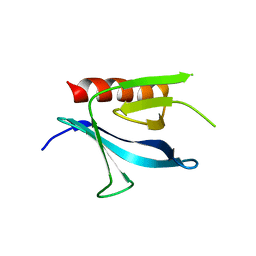 | | Crystal Structure of the Carboxyl Terminal PH Domain of Pleckstrin To 2.1 Angstroms | | Descriptor: | Pleckstrin | | Authors: | Jackson, S.G, Zhang, Y, Zhang, K, Summerfield, R, Haslam, R.J, Junop, M.S. | | Deposit date: | 2005-05-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the carboxy-terminal PH domain of pleckstrin at 2.1 Angstroms.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ANO
 
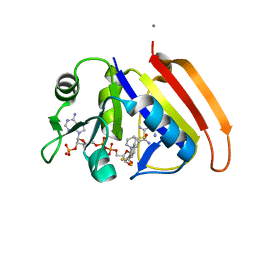 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
7RUE
 
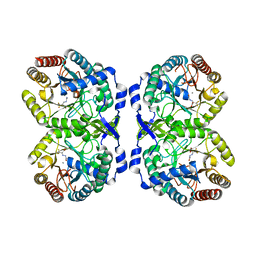 | | DAHP synthase complexed with trifluoropyruvate semicarbazone | | Descriptor: | (2E)-2-(2-carbamoylhydrazinylidene)-3,3,3-trifluoropropanoic acid, MANGANESE (II) ION, Phospho-2-dehydro-3-deoxyheptonate aldolase, ... | | Authors: | Heimhalt, M, Mukherjee, P, Grainger, R, Szabla, R, Brown, C, Turner, R, Junop, M.S, Berti, P.J. | | Deposit date: | 2021-08-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Inhibitor-in-Pieces Approach to DAHP Synthase Inhibition: Potent Enzyme and Bacterial Growth Inhibition.
Acs Infect Dis., 7, 2021
|
|
2ANQ
 
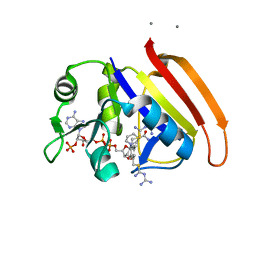 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
7RUD
 
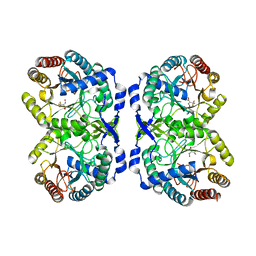 | | DAHP synthase complex with trifluoropyruvate oxime | | Descriptor: | (2Z)-3,3,3-trifluoro-2-(hydroxyimino)propanoic acid, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive | | Authors: | Heimhalt, M, Mukherjee, P, Grainger, R, Szabla, R, Brown, C, Turner, R, Junop, M.S, Berti, P.J. | | Deposit date: | 2021-08-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Inhibitor-in-Pieces Approach to DAHP Synthase Inhibition: Potent Enzyme and Bacterial Growth Inhibition.
Acs Infect Dis., 7, 2021
|
|
5KUE
 
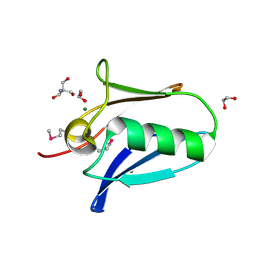 | | Human SeMet incorporated I141M/L146M mitochondrial calcium uniporter (residues 72-189) crystal structure with magnesium | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Calcium uniporter protein, ... | | Authors: | Mok, C.Y.M, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
5KUI
 
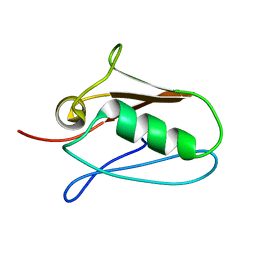 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with calcium. | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
5KUJ
 
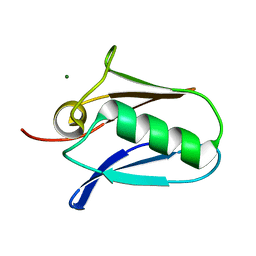 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with magnesium. | | Descriptor: | Calcium uniporter protein, mitochondrial, MAGNESIUM ION | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
5KUG
 
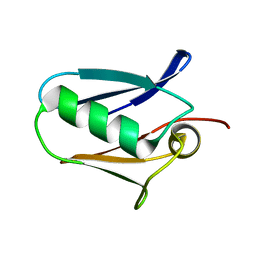 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with lithium | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
1EA6
 
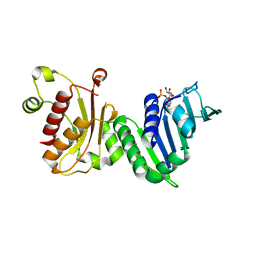 | |
