2MBV
 
 | | LMO4-LIM2 in complex with DEAF1 (404-418) | | Descriptor: | Fusion protein of LIM domain transcription factor LMO4 (77-147) and DEAF1 (404-418), ZINC ION | | Authors: | Joseph, S, Matthews, J.M, Kwan, A.H.-Y, Mackay, J.P, Cubeddu, L, Foo, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-20 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of LIM-only protein 4 (LMO4) in complex with Deformed Epidermal Autoregulatory Factor-1 (DEAF1)
To be Published
|
|
6U0D
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
6UWU
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
2GV3
 
 | | Translocation of a tRNA with an extended anticodon through the ribosome | | Descriptor: | 5'-R(*GP*GP*CP*CP*AP*GP*AP*CP*UP*CP*CP*CP*GP*AP*AP*UP*CP*UP*GP*GP*CP*C)-3' | | Authors: | Phelps, S.S, Gaudin, C, Yoshizawa, S, Benitez, C, Fourmy, D, Joseph, S. | | Deposit date: | 2006-05-02 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Translocation of a tRNA with an Extended Anticodon Through the Ribosome.
J.Mol.Biol., 360, 2006
|
|
2UXD
 
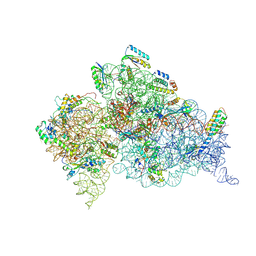 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXC
 
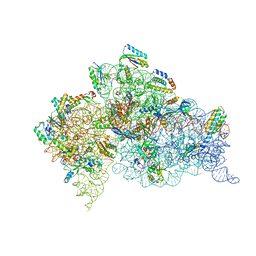 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
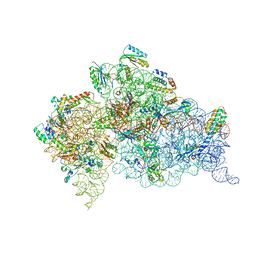 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
7UI6
 
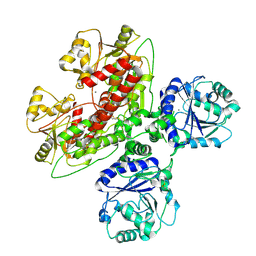 | |
4ZW2
 
 | |
3QQU
 
 | | Cocrystal structure of unphosphorylated igf with pyrimidine 8 | | Descriptor: | Insulin-like growth factor 1 receptor, N~2~-[3-methoxy-4-(morpholin-4-yl)phenyl]-N~4~-(quinolin-3-yl)pyrimidine-2,4-diamine | | Authors: | Huang, X. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1I2U
 
 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
6WU8
 
 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | Descriptor: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Joseph, S, Rodenberger, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
2FK8
 
 | |
2FK7
 
 | |
7UI7
 
 | |
