6P0U
 
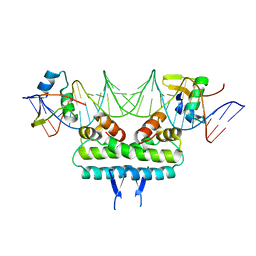 | | Crystal structure of ternary DNA complex " FX(1-2)-2Xis" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
6P0S
 
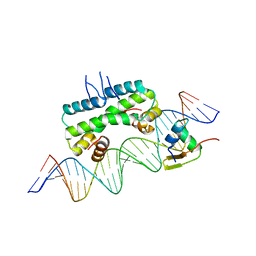 | | Crystal structure of ternary DNA complex "FX2" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
6P0T
 
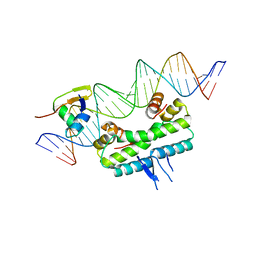 | | Crystal structure of ternary DNA complex "FX(1-2)-1Xis" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
5DTD
 
 | |
5E3L
 
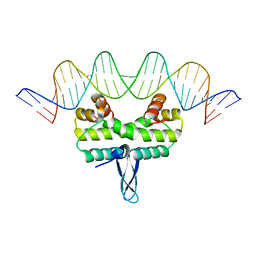 | |
5DS9
 
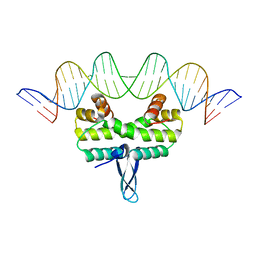 | |
5E3O
 
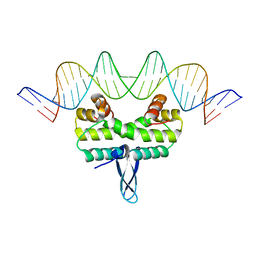 | |
5E3N
 
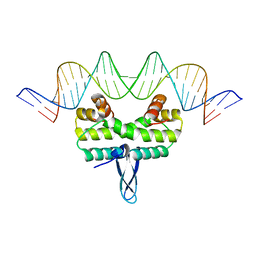 | |
5E3M
 
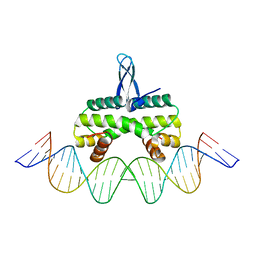 | | Crystal structure of Fis bound to 27bp DNA F35 (AAATTAGTTTGAATCTCGAGCTAATTT) | | Descriptor: | DNA (27-MER), DNA-binding protein Fis | | Authors: | Stella, S, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2015-10-03 | | Release date: | 2016-03-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.
Plos One, 11, 2016
|
|
3FIS
 
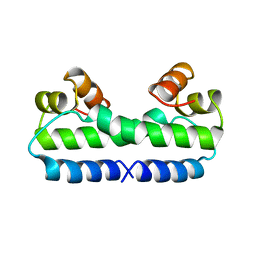 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6DGB
 
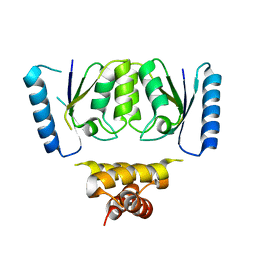 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
6DGC
 
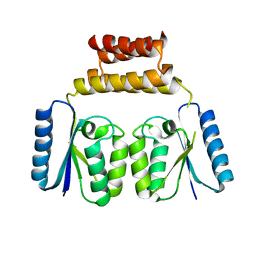 | | Crystal structure of the C-terminal catalytic domain of ISC1926 TnpA, an IS607-like serine recombinase | | Descriptor: | ISC1926 TnpA C-terminal catalytic domain | | Authors: | Hancock, S.P, Kumar, P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
1HCR
 
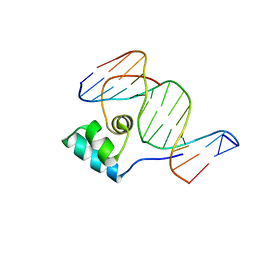 | |
4FIS
 
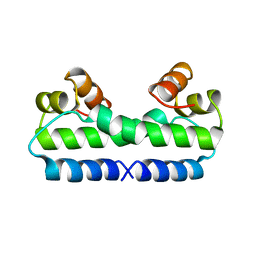 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J.-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1IJW
 
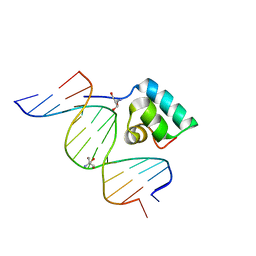 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*(CBR)P*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-04-30 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JJ8
 
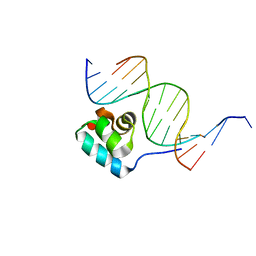 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*(5IT)P*TP*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', DNA-INVERTASE HIN | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-03 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JJ6
 
 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*(5IT)P*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-03 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKR
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*CP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKO
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*CP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*GP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKP
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*CP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*GP*AP*AP*GP*A)-3', DNA-INVERTASE HIN | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKQ
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*AP*TP*AP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3', DNA-INVERTASE HIN | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
4IHW
 
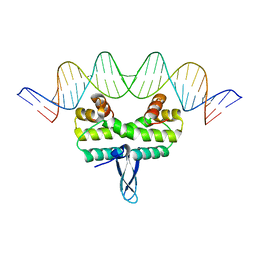 | |
4IHY
 
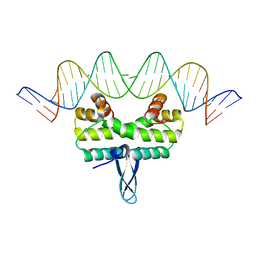 | |
4IHV
 
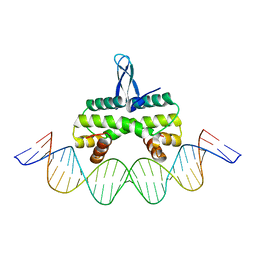 | |
4IHX
 
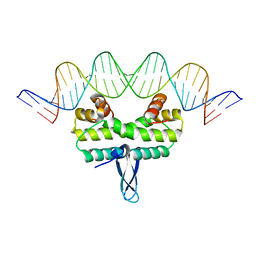 | |
