4V8O
 
 | | Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3 | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-07-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Hybrid State of Ribosome in Complex with the Guanosine Triphosphatase Release Factor 3.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V5J
 
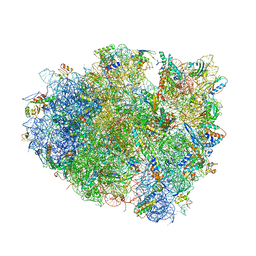 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | Descriptor: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | Deposit date: | 2010-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2VXW
 
 | |
3S2Y
 
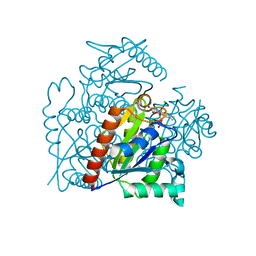 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
2PCW
 
 | |
2PCV
 
 | |
5Y7Q
 
 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
5Z6W
 
 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
6J3Q
 
 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
5Y7G
 
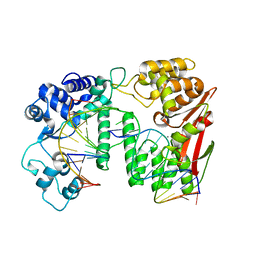 | | Crystal structure of paFAN1 bound to 1nt 5'flap DNA with gap | | Descriptor: | CALCIUM ION, DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Jin, H. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
6JPK
 
 | |
4V5E
 
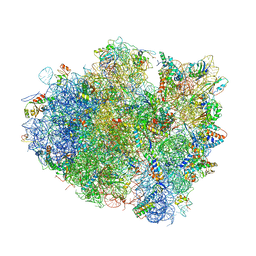 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
7RR5
 
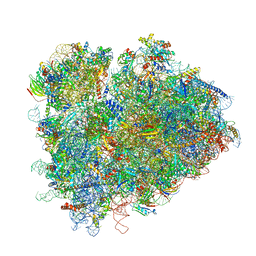 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
5U4I
 
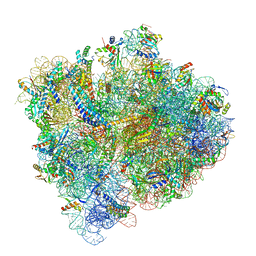 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5U4J
 
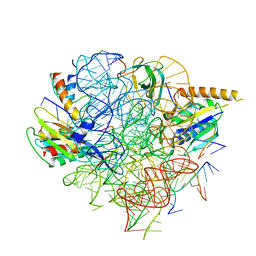 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5GVV
 
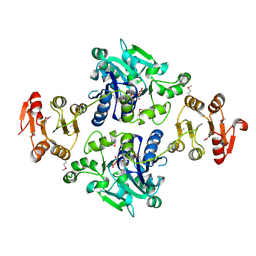 | | Crystal structure of the glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5GVW
 
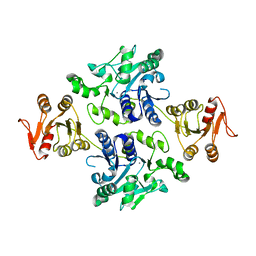 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
3TEQ
 
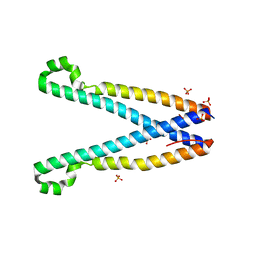 | | Crystal structure of SOAR domain | | Descriptor: | PHOSPHATE ION, Stromal interaction molecule 1 | | Authors: | Yang, X, Jin, H, Cai, X, Shen, Y. | | Deposit date: | 2011-08-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1).
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TER
 
 | |
8GQW
 
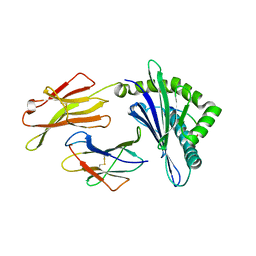 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | Hu64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
8GQV
 
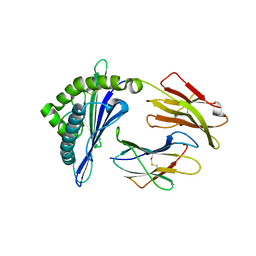 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | As64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
6C4I
 
 | |
6C4H
 
 | |
6C5L
 
 | |
6KHE
 
 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
