1BVB
 
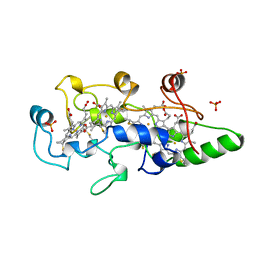 | | HEME-PACKING MOTIFS REVEALED BY THE CRYSTAL STRUCTURE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C-554, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Iverson, T.M, Arciero, D.M, Hsu, B.T, Logan, M.S.P, Hooper, A.B, Rees, D.C. | | Deposit date: | 1998-09-16 | | Release date: | 1999-05-18 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea.
Nat.Struct.Biol., 5, 1998
|
|
1FT5
 
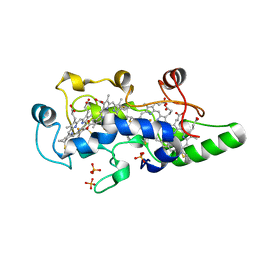 | | CRYSTAL STRUCTURE OF THE OXIDIZED STATE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C554, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Iverson, T.M, Arciero, D.M, Hooper, A.B, Rees, D.C. | | Deposit date: | 2000-09-11 | | Release date: | 2000-09-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea.
J.Biol.Inorg.Chem., 6, 2001
|
|
1FT6
 
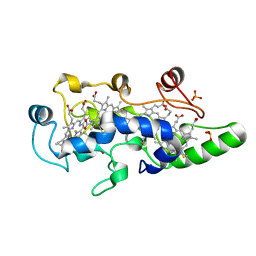 | | REDUCED STATE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C554, DITHIONITE, HEME C, ... | | Authors: | Iverson, T.M, Arciero, D.M, Hooper, A.B, Rees, D.C. | | Deposit date: | 2000-09-11 | | Release date: | 2000-09-20 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea.
J.Biol.Inorg.Chem., 6, 2001
|
|
1L0V
 
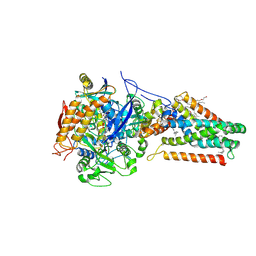 | | Quinol-Fumarate Reductase with Menaquinol Molecules | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1KFY
 
 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1KF6
 
 | | E. coli Quinol-Fumarate Reductase with Bound Inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-19 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1QRE
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRL
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRF
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION, SULFATE ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRM
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, SULFATE ION, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QQ0
 
 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-10 | | Release date: | 1999-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRG
 
 | | A CLOSER LOOK AND THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
3UN3
 
 | | phosphopentomutase T85Q variant soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UO0
 
 | | phosphorylated Bacillus cereus phosphopentomutase soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UN5
 
 | | Bacillus cereus phosphopentomutase T85E variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UN2
 
 | | Phosphopentomutase T85Q variant enzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UNY
 
 | | Bacillus cereus phosphopentomutase T85E variant soaked with glucose 1,6-bisphosphate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
6AWF
 
 | | Escherichia coli quinol:fumarate reductase crystallized without dicarboxylate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | New crystal forms of the integral membrane Escherichia coli quinol:fumarate reductase suggest that ligands control domain movement.
J. Struct. Biol., 202, 2018
|
|
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFI
 
 | | SK678 binding region (Siglec + Unique) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFB
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, GLYCEROL, SK150 siglec + Unique, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EF7
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFA
 
 | | GspB Siglec + Unique domains | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EF9
 
 | | GspB Siglec domain | | Descriptor: | FORMIC ACID, Platelet binding protein GspB - Siglec domain, SODIUM ION | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFD
 
 | | Hsa Siglec and Unique domains in complex with the sialyl T antigen trisaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
