6ZYC
 
 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZXP
 
 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
4YGM
 
 | | Vaccinia virus his-D4/A20(1-50) in complex with uracil | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, URACIL, ... | | Authors: | Tarbouriech, N, Iseni, F, Burmeister, W.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
4YIG
 
 | | vaccinia virus D4/A20(1-50) in complex with dsDNA containing an abasic site and free uracyl | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(ORP)P*AP*TP*CP*TP*T)-3'), DNA polymerase processivity factor component A20, ... | | Authors: | tarbouriech, N, burmeister, W.P, iseni, F. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
4ODA
 
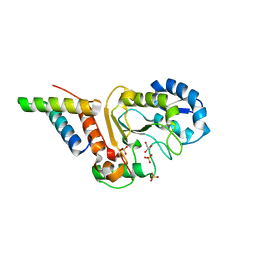 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
4OD8
 
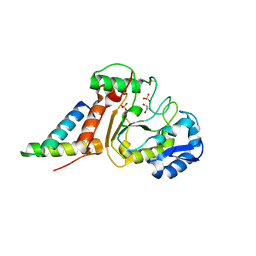 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
5JKT
 
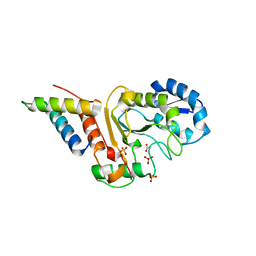 | | vaccinia virus D4 P173G mutant /A20(1-50) | | Descriptor: | ACETATE ION, DNA polymerase processivity factor component A20, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Peyrefitte, C.N, Iseni, F. | | Deposit date: | 2016-04-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural analysis of point mutations at the Vaccinia virus A20/D4 interface.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JKS
 
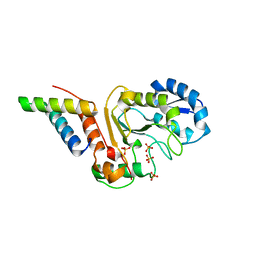 | | vaccinia virus D4 R167A mutant /A20(1-50) | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Peyrefitte, C.N, Iseni, F. | | Deposit date: | 2016-04-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analysis of point mutations at the Vaccinia virus A20/D4 interface.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JKR
 
 | | vaccinia virus D4/A20(1-50)w43a mutant | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Peyrefitte, C.N, Iseni, F. | | Deposit date: | 2016-04-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of point mutations at the Vaccinia virus A20/D4 interface.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5N2E
 
 | | Structure of the E9 DNA polymerase from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
5N2H
 
 | | Structure of the E9 DNA polymerase exonuclease deficient mutant (D166A+E168A) from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
5N2G
 
 | | Structure of the E9 DNA polymerase from vaccinia virus in complex with manganese | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
6QNT
 
 | |
6QNU
 
 | |
