8HTA
 
 | |
5X7Y
 
 | | Crystal Structure of the Dog Lipocalin Allergen Can f 6 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipocalin-Can f 6 allergen | | Authors: | Yamamoto, K, Otani, T, Sugiura, K, Nakatsuji, M, Nishimura, S, Inui, T. | | Deposit date: | 2017-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the dog allergen Can f 6 and structure-based implications of its cross-reactivity with the cat allergen Fel d 4.
Sci Rep, 9, 2019
|
|
2RQ0
 
 | |
1MKC
 
 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
6LK4
 
 | | Crystal structure of GMP reductase from Trypanosoma brucei in complex with guanosine 5'-triphosphate | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Guanosine 5'-monophosphate Reductase, PHOSPHATE ION | | Authors: | Mase, H, Otani, T, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6JIG
 
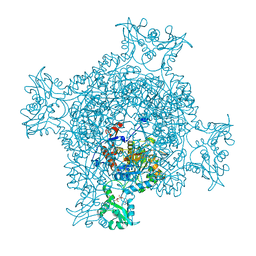 | | Crystal structure of GMP reductase C318A from Trypanosoma brucei in complex with guanosine 5'-monophosphate | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Mase, H, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-02-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6JL8
 
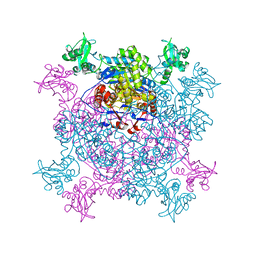 | |
2L87
 
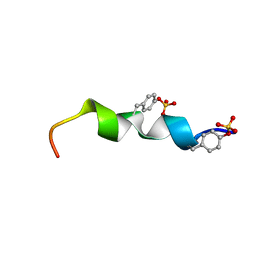 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | Deposit date: | 2011-01-06 | | Release date: | 2011-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
7DRU
 
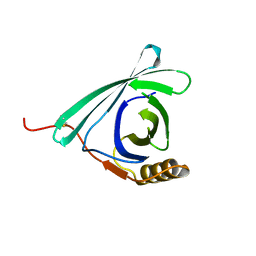 | |
3AFB
 
 | |
3VOC
 
 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
2KTD
 
 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | Descriptor: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | Authors: | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
2LAA
 
 | |
2LAB
 
 | |
