2DJI
 
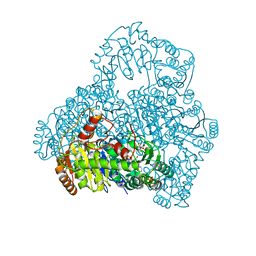 | | Crystal Structure of Pyruvate Oxidase from Aerococcus viridans containing FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate oxidase, SULFATE ION | | Authors: | Juan, E.C.M, Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3VW8
 
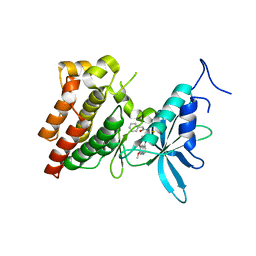 | | Crystal structure of human c-Met kinase domain with its inhibitor | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-({4-[(6,7-dimethoxyquinolin-4-yl)oxy]phenyl}carbamothioyl)-2-phenylacetamide | | Authors: | Matsumoto, S, Miyamoto, N, Hirayama, T, Oki, H, Okada, K, Tawada, M, Iwata, H, Miki, H, Nakamura, K, Hori, A, Imamura, S. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design, synthesis, and evaluation of imidazo[1,2-b]pyridazine and imidazo[1,2-a]pyridine derivatives as novel dual c-Met and VEGFR2 kinase inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
3VDR
 
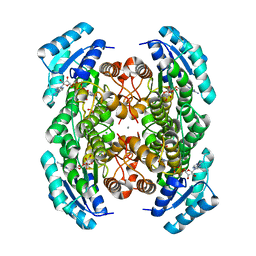 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1VAX
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis | | Descriptor: | Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter globiformis
To be Published
|
|
1V5G
 
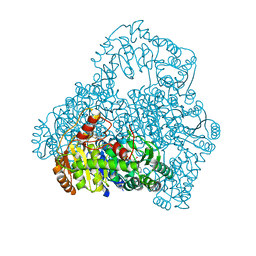 | | Crystal Structure of the Reaction Intermediate between Pyruvate oxidase containing FAD and TPP, and Substrate Pyruvate | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1V5F
 
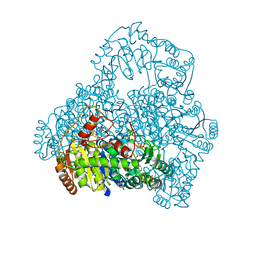 | | Crystal Structure of Pyruvate oxidase complexed with FAD and TPP, from Aerococcus viridans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate oxidase, ... | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1VAY
 
 | | Crystal Structure of Uricase from Arthrobacter globiformis with inhibitor 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, Uric acid oxidase | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2004-02-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Uricase from Arthrobacter Globiformis Complexed with an inhibitor 8-Azaxanthine
To be Published
|
|
1V5E
 
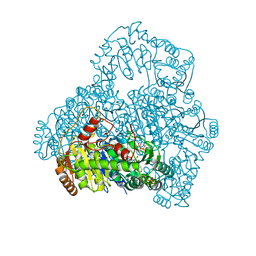 | | Crystal Structure of Pyruvate oxidase containing FAD, from Aerococcus viridans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate oxidase, SULFATE ION | | Authors: | Hossain, M.T, Suzuki, K, Yamamoto, T, Imamura, S, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Pyruvate oxidase containing FAD, from Aerococcus viridans
To be Published
|
|
3VHK
 
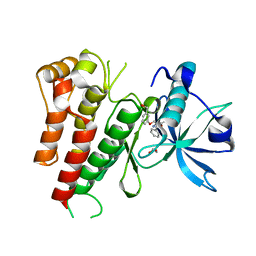 | | Crystal structure of the VEGFR2 kinase domain in complex with a back pocket binder | | Descriptor: | 1,2-ETHANEDIOL, Vascular endothelial growth factor receptor 2, {3-[(5-methyl-2-phenyl-1,3-oxazol-4-yl)methoxy]phenyl}methanol | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
3VID
 
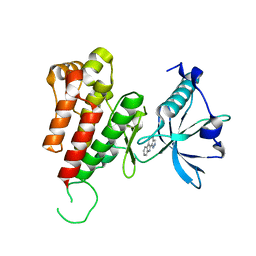 | | Crystal structure of human VEGFR2 kinase domain with Compound A. | | Descriptor: | 4,5,6,11-tetrahydro-1H-pyrazolo[4',3':6,7]cyclohepta[1,2-b]indole, Vascular endothelial growth factor receptor 2 | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-09-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
6P4V
 
 | |
6PGR
 
 | |
6WL6
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6WKZ
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
2YZ7
 
 | | X-ray analyses of 3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase | | Authors: | Hoque, M.M, Juan, E.C.M, Shimizu, S, Hossain, M.T, Takenaka, A. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structures of Alcaligenes faecalisD-3-hydroxybutyrate dehydrogenase before and after NAD(+) and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4HY5
 
 | | Crystal structure of cIAP1 BIR3 bound to T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Snell, G.P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4HY4
 
 | | Crystal structure of cIAP1 BIR3 bound to T3170284 | | Descriptor: | (3S,8aR)-2-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4HY0
 
 | | Crystal structure of XIAP BIR3 with T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Snell, G.S, Dougan, D.R. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
1LWB
 
 | |
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1IT5
 
 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
3VDQ
 
 | | Crystal structure of alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase in complex with NAD(+) and acetate | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hoque, M.M, Shimizu, S, Hossain, M.T, Yamamoto, T, Suzuki, K, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of Alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase before and after NAD+ and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VO3
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with imidazo[1,2-b]pyridazine derivative | | Descriptor: | 1,2-ETHANEDIOL, N-[3-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)phenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H, Okada, K. | | Deposit date: | 2012-01-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-[5-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)-2-methylphenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide (TAK-593), a highly potent VEGFR2 kinase inhibitor
Bioorg.Med.Chem., 21, 2013
|
|
3VHE
 
 | | Crystal structure of human VEGFR2 kinase domain with a novel pyrrolopyrimidine inhibitor. | | Descriptor: | 1-{2-fluoro-4-[(5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Vascular endothelial growth factor receptor 2 | | Authors: | Oguro, Y, Miyamoto, N, Okada, K, Takagi, T, Iwata, H, Awazu, Y, Miki, H, Hori, A, Kamiyama, K, Imanura, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, synthesis, and evaluation of 5-methyl-4-phenoxy-5H-pyrrolo[3,2-d]pyrimidine derivatives: novel VEGFR2 kinase inhibitors binding to inactive kinase conformation.
Bioorg.Med.Chem., 18, 2010
|
|
2YZE
 
 | | Crystal structure of uricase from Arthrobacter globiformis | | Descriptor: | (dihydroxyboranyloxy-hydroxy-boranyl)oxylithium, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
