1A05
 
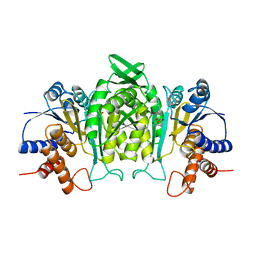 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
5B0O
 
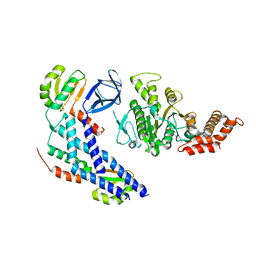 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1IPD
 
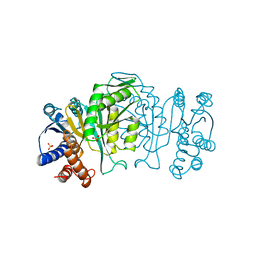 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
2D4V
 
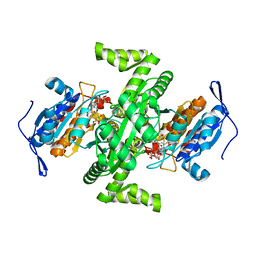 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | Descriptor: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | Authors: | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
2D4W
 
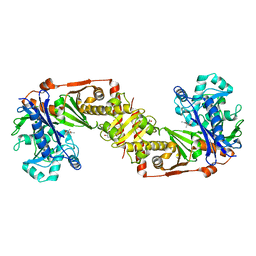 | |
2D4Y
 
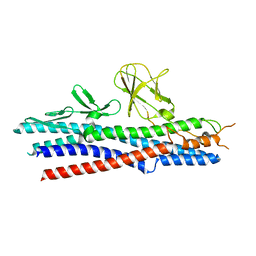 | |
2D4X
 
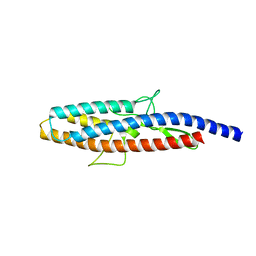 | |
2DPY
 
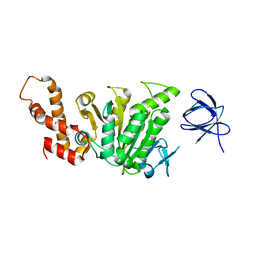 | |
3A7M
 
 | | Structure of FliT, the flagellar type III chaperone for FliD | | Descriptor: | Flagellar protein fliT | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5I
 
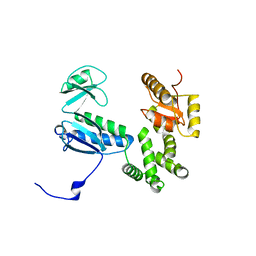 | |
3AJW
 
 | | Structure of FliJ, a soluble component of flagellar type III export apparatus | | Descriptor: | Flagellar fliJ protein, MERCURY (II) ION | | Authors: | Imada, K, Ibuki, T, Minamino, T, Namba, K. | | Deposit date: | 2010-06-23 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases
Nat.Struct.Mol.Biol., 18, 2011
|
|
2D4U
 
 | | Crystal Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr | | Descriptor: | Methyl-accepting chemotaxis protein I | | Authors: | Imada, K, Tajima, H, Namba, K, Sakuma, M, Homma, M, Kawagishi, I. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in the bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 2011
|
|
3W0E
 
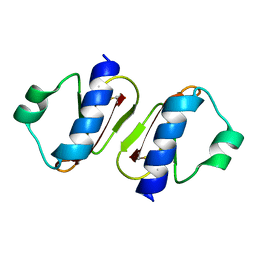 | | Structure of elastase inhibitor AFUEI (crystal form II) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
3W0D
 
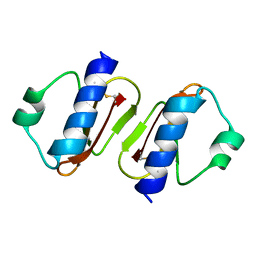 | | Structure of elastase inhibitor AFUEI (cyrstal form I) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
2ZVZ
 
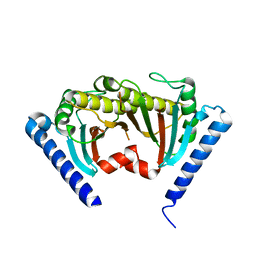 | |
3W1C
 
 | |
3W1D
 
 | |
2ZF8
 
 | | Crystal structure of MotY | | Descriptor: | Component of sodium-driven polar flagellar motor | | Authors: | Imada, K, Kojima, S, Namba, K, Homma, M. | | Deposit date: | 2007-12-25 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZOV
 
 | |
3AJC
 
 | | Structure of the MC domain of FliG (PEV), a CW-biased mutant | | Descriptor: | Flagellar motor switch protein fliG | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rotational switching mechanism of the bacterial flagellar motor
Plos Biol., 9, 2011
|
|
3ADY
 
 | | Crystal structure of DotD from Legionella | | Descriptor: | DotD | | Authors: | Imada, K, Nakano, N, Kubori, T, Kinoshita, M, Nagai, H. | | Deposit date: | 2010-01-29 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems
Plos Pathog., 6, 2010
|
|
5H72
 
 | | Structure of the periplasmic domain of FliP | | Descriptor: | Flagellar biosynthetic protein FliP | | Authors: | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-11-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
8GQY
 
 | | CryoEM structure of pentameric MotA from Aquifex aeolicus | | Descriptor: | Motility protein A | | Authors: | Nishikino, T, Takekawa, N, Kishikawa, J, Hirose, M, Onoe, S, Kato, T, Imada, K. | | Deposit date: | 2022-08-31 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of MotA, a flagellar stator protein, from hyperthermophile.
Biochem.Biophys.Res.Commun., 631, 2022
|
|
1VGL
 
 | | Crystal structure of tetrameric KaiB from T.elongatus BP-1 | | Descriptor: | Circadian clock protein kaiB, MERCURY (II) ION | | Authors: | Iwase, R, Imada, K, Hayashi, F, Uzumaki, T, Namba, K, Ishiura, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex.
J.Biol.Chem., 280, 2005
|
|
8I4J
 
 | |
