1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1L1J
 
 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | Descriptor: | heat shock protease HtrA | | Authors: | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2002-02-18 | | Release date: | 2003-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
4YGQ
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
3QBQ
 
 | | Crystal structure of extracellular domains of mouse RANK-RANKL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 11, Tumor necrosis factor receptor superfamily member 11A | | Authors: | Ta, H.M, Nguyen, G.T.T, Jin, H.M, Choi, J.K, Park, H, Kim, N.S, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LFU
 
 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | Descriptor: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LGW
 
 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | Descriptor: | GLYCEROL, Regulatory protein SdiA | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5LS0
 
 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | Authors: | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
3F21
 
 | | Crystal structure of Zalpha in complex with d(CACGTG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DAP*DCP*DGP*DTP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3EYI
 
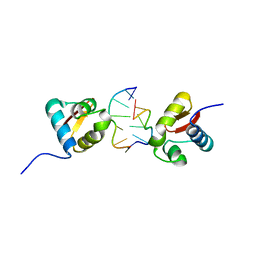 | |
3F23
 
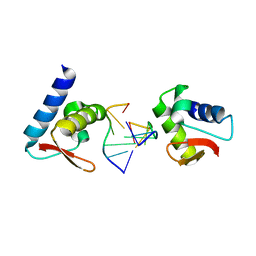 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F22
 
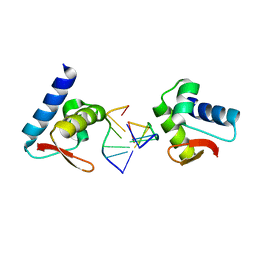 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3SH4
 
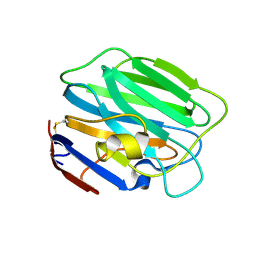 | |
3SH5
 
 | |
4KIA
 
 | | Crystal structure of LmHde, heme-degrading enzyme, from Listeria monocytogenes | | Descriptor: | Lmo2213 protein | | Authors: | Kim, K.K, Duong, T, Kim, T. | | Deposit date: | 2013-05-02 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional characterization of an Isd-type haem-degradation enzyme from Listeria monocytogenes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KMF
 
 | |
