4R9V
 
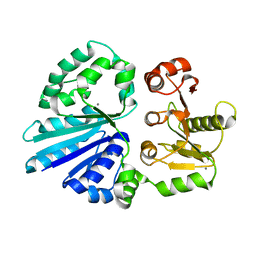 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R84
 
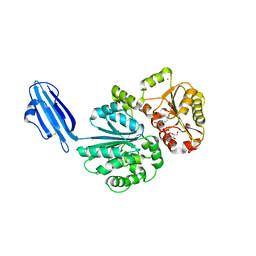 | | Crystal structure of Sialyltransferase from Photobacterium damsela with CMP-3F(a)Neu5Ac bound | | Descriptor: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4IME
 
 | |
4IMG
 
 | |
4IMC
 
 | |
4IMD
 
 | |
4IMF
 
 | |
4R83
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
3BEG
 
 | | Crystal structure of SR protein kinase 1 complexed to its substrate ASF/SF2 | | Descriptor: | ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PHOSPHOSERINE, ... | | Authors: | Ngo, J.C, Giang, K, Chakrabarti, S, Ma, C.-T, Huynh, N, Hagopian, J, Dorrestein, P.C, Fu, X.-D, Adams, J.A, Ghosh, G. | | Deposit date: | 2007-11-18 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A sliding docking interaction is essential for sequential and
processive phosphorylation of an SR protein by SRPK1
Mol.Cell, 29, 2008
|
|
5FMO
 
 | | Crystal structure and proteomics analysis of empty virus like particles of Cowpea mosaic virus | | Descriptor: | EMPTY VIRUS LIKE PARTICLES OF COWPEA MOSAIC VIRUS | | Authors: | Huynh, N, Hesketh, E.L, Saxena, P, Meshcheriakova, Y, Ku, Y.C, Hoang, L, Johnson, J.E, Ranson, N.A, Lomonossoff, G.P, Reddy, V.S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Proteomics Analysis of Empty Virus-Like Particles of Cowpea Mosaic Virus.
Structure, 24, 2016
|
|
8CX9
 
 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
8E17
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6M)-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-4-[5-(methanesulfonyl)-2-methoxyphenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-09 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8DYR
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E3W
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
5ZKA
 
 | | Crystal structure of N-acetylneuraminate lyase from Fusobacterium nucleatum complexed with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, N-acetylneuraminate lyase, TRIETHYLENE GLYCOL | | Authors: | Kumar, J.P, Rao, H, Nayak, V, Ramaswamy, S. | | Deposit date: | 2018-03-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
