2NQP
 
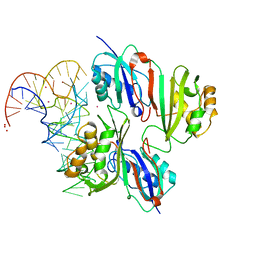 | |
2NRE
 
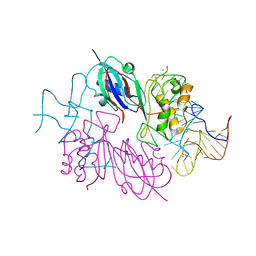 | |
2NR0
 
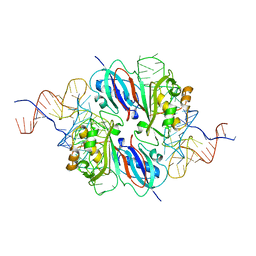 | |
8SRO
 
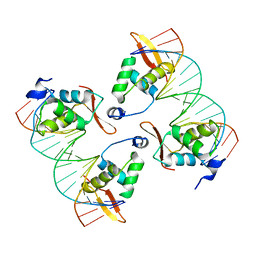 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
8SRP
 
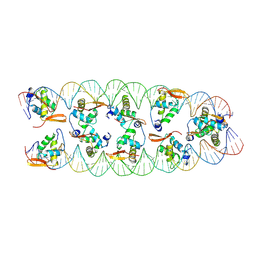 | |
7TDW
 
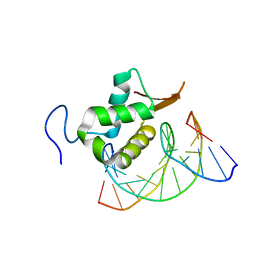 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
7TDX
 
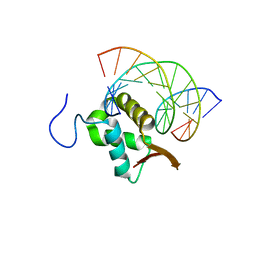 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
3J6J
 
 | | 3.6 Angstrom resolution MAVS filament generated from helical reconstruction | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Wu, B, Peisley, A, Li, Z, Egelman, E, Walz, T, Penczek, P, Hur, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I.
Mol.Cell, 55, 2014
|
|
7JL3
 
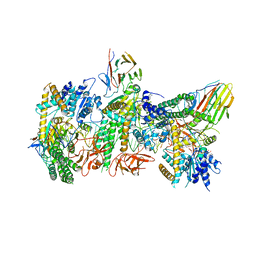 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
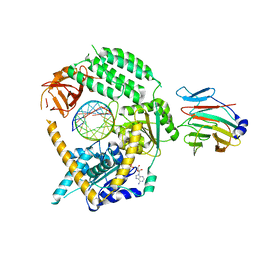 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
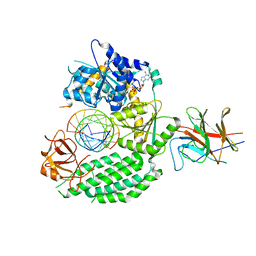 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL4
 
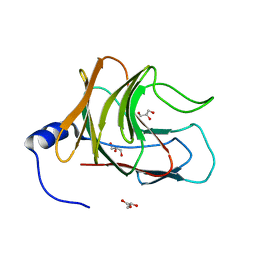 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL2
 
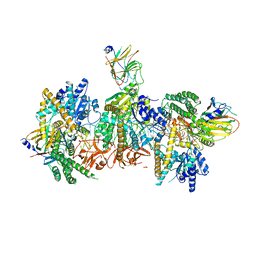 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
4GL2
 
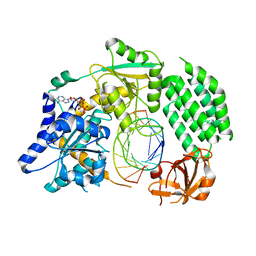 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
4NQK
 
 | | Structure of an Ubiquitin complex | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, Ubiquitin | | Authors: | Peisley, A, Wu, B, Hur, S. | | Deposit date: | 2013-11-25 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I.
Nature, 509, 2014
|
|
4P4H
 
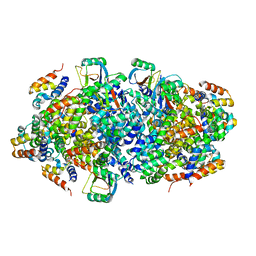 | |
4MSS
 
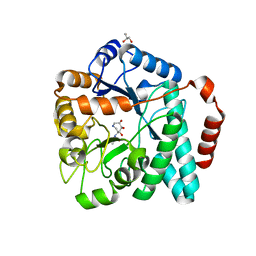 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
