3G3X
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 100 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3V
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
1ZYT
 
 | | Crystal structure of spin labeled T4 Lysozyme (A82R1) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | EPR (1.7 Å), X-RAY DIFFRACTION | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins
Protein Sci., 18, 2009
|
|
2CUU
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-05-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins
Protein Sci., 18, 2009
|
|
2IGC
 
 | | Structure of Spin labeled T4 Lysozyme Mutant T115R1A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-09-22 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
1ZUR
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1F) | | Descriptor: | CHLORIDE ION, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-4-phenyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-05-31 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1F)
To be Published
|
|
1ZWN
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1B) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-03 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1B)
To be Published
|
|
2A4T
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R7) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-29 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R7
To be Published
|
|
3HWL
 
 | | Crystal Structure of T4 lysozyme with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 131 | | Descriptor: | AZIDE ION, CHLORIDE ION, Lysozyme | | Authors: | Fleissner, M.R, Cascio, D, Schultz, P.G, Hubbell, W.L. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed spin labeling of a genetically encoded unnatural amino acid.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K2R
 
 | | Crystal Structure of Spin Labeled T4 Lysozyme Mutant K65V1/R76V1 | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, Lysozyme, ... | | Authors: | Toledo Warshaviak, D, Cascio, D, Khramtsov, V.V, Hubbell, W.L. | | Deposit date: | 2009-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Spin Labeled T4 Lysozyme Mutant K65V1/R76V1
To be Published
|
|
3L2X
 
 | |
5W0P
 
 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
2NTH
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant L118R1 | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2NTG
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OU8
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1 at Room Temperature | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | EPR (1.8 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OU9
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2Q9D
 
 | | Structure of spin-labeled T4 lysozyme mutant A41R1 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (1.4 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2Q9E
 
 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
4BUM
 
 | |
5D4N
 
 | | Structure of CPII bound to ADP, AMP and acetate, from Thiomonas intermedia K12 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5DRK
 
 | | 2.3 Angstrom Structure of CPII, a nitrogen regulatory PII-like protein from Thiomonas intermedia K12, bound to ADP, AMP and bicarbonate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-09-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5DS7
 
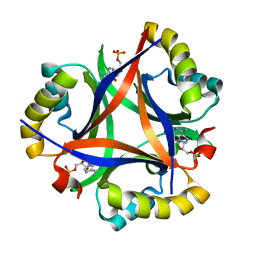 | | 2.0 A Structure of CPII, a nitrogen regulatory PII-like protein from Thiomonas intermedia K12, bound AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Nitrogen regulatory protein P-II | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-09-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5F1S
 
 | |
