1NHH
 
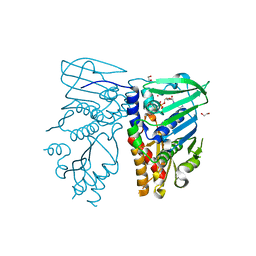 | | Crystal structure of N-terminal 40KD MutL protein (LN40) complex with ADPnP and one Rubidium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1NHJ
 
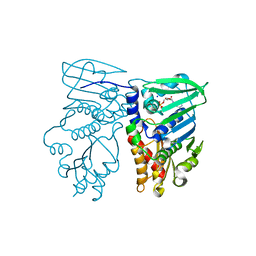 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | Descriptor: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1NHI
 
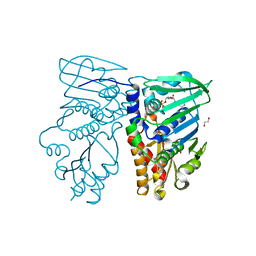 | | Crystal structure of N-terminal 40KD MutL (LN40) complex with ADPnP and one potassium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
8Y33
 
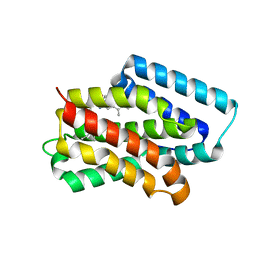 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 2024
|
|
3B83
 
 | |
3LA1
 
 | |
3UUG
 
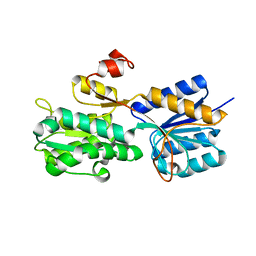 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3URM
 
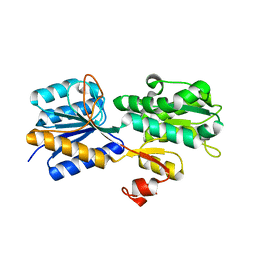 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2RBL
 
 | |
2RB8
 
 | |
8J1W
 
 | |
8J1X
 
 | |
3RPL
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
3ROJ
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
8I8Y
 
 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8J98
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo backbone design for monomerization of near-infrared fluorescent protein
To Be Published
|
|
8JPA
 
 | |
3GEX
 
 | |
3I19
 
 | |
6U7V
 
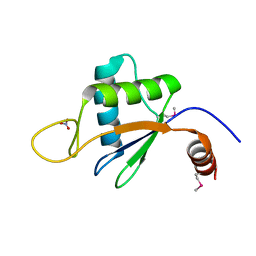 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
8HDU
 
 | |
8HDV
 
 | |
2G6P
 
 | | Crystal structure of truncated (delta 1-89) human methionine aminopeptidase Type 1 in complex with Pyridyl pyrimidine derivative | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-CHLORO-6-METHYL-N-(2-PHENYLETHYL)-2-PYRIDIN-2-YLPYRIMIDIN-4-AMINE, COBALT (II) ION, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2006-02-24 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Pyridinylpyrimidines as Inhibitors of Human Methionine Aminopeptidases.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2XWT
 
 | | CRYSTAL STRUCTURE OF THE TSH RECEPTOR IN COMPLEX WITH A BLOCKING TYPE TSHR AUTOANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, THYROID BLOCKING HUMAN AUTOANTIBODY K1-70 HEAVY CHAIN, ... | | Authors: | Sanders, J, Sanders, P, Young, S, Kabelis, K, Baker, S, Sullivan, A, Evans, M, Clark, J, Wilmot, J, Hu, X, Roberts, E, Powell, M, Nunez Miguel, R, Furmaniak, J, Rees Smith, B. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
J.Mol.Endocrinol., 46, 2011
|
|
6S8Y
 
 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
