1VAQ
 
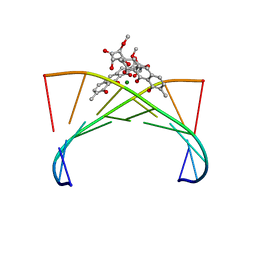 | | Crystal structure of the Mg2+-(chromomycin A3)2-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by metal ion | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Robinson, H, Gao, Y.G, Wang, A.H.-J. | | Deposit date: | 2004-02-19 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion
Nucleic Acids Res., 32, 2004
|
|
5ZT2
 
 | |
5DEV
 
 | |
5YTZ
 
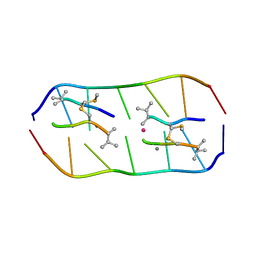 | | Crystal structure of echinomycin-d(ACGTCGT)2 complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*TP*CP*GP*(UD))-3'), Echinomycin, ... | | Authors: | Hou, M.H, Wu, P.C, Kao, Y.F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cooperative recognition of T:T mismatch by echinomycin causes structural distortions in DNA duplex
Nucleic Acids Res., 46, 2018
|
|
5YZE
 
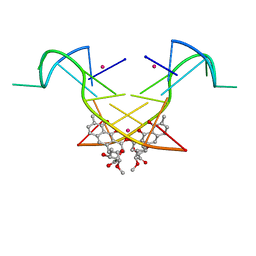 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
5YTY
 
 | | Crystal structure of echinomycin-d(ACGACGT/ACGTCGT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*AP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Wu, P.C, Kao, Y.F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Cooperative recognition of T:T mismatch by echinomycin causes structural distortions in DNA duplex
Nucleic Acids Res., 46, 2018
|
|
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
 | |
6L75
 
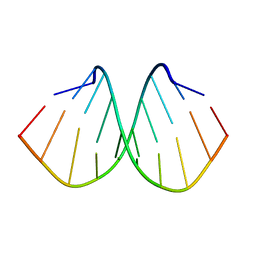 | | Crystal structure of d(GTGGGCCGAC)2 DNA duplex | | Descriptor: | DNA (5'-D(P*GP*TP*GP*GP*GP*CP*CP*GP*AP*C)-3') | | Authors: | Hou, M.H, Satange, R.B, Zeng, J.Y. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6L76
 
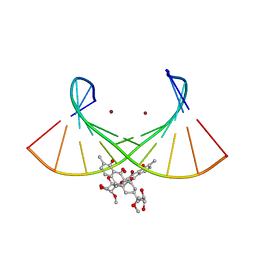 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6M5J
 
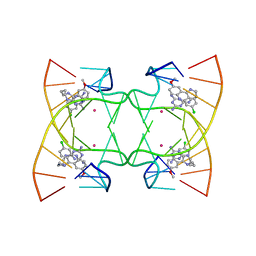 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA/TTCTGCAGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*TP*TP*CP*TP*GP*CP*AP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6M4T
 
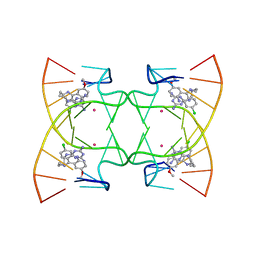 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
6LNN
 
 | |
6LZ6
 
 | |
6LZ8
 
 | |
7DYD
 
 | |
5GUN
 
 | | Crystal structure of d(GTGGAATGGAAC) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*TP*GP*GP*AP*AP*TP*GP*GP*AP*AP*C)-3') | | Authors: | Satange, R.B, Kao, Y.F, Hou, M.H. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LM7
 
 | |
4PZQ
 
 | | Crystal Structure of CCG DNA repeats | | Descriptor: | COBALT (II) ION, DNA (5'-D(*TP*CP*CP*GP*CP*CP*GP*CP*CP*GP*A)-3') | | Authors: | Chen, Y.W, Hou, M.H. | | Deposit date: | 2014-03-31 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for the identification of an i-motif tetraplex core with a parallel-duplex junction as a structural motif in CCG triplet repeats
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4QNN
 
 | |
8IQJ
 
 | | Crystal structure of SARS-CoV2 N-NTD | | Descriptor: | Nucleoprotein | | Authors: | Hong, J.Y, Hou, M.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2.
Biophys.J., 123, 2024
|
|
8IV3
 
 | |
8J6X
 
 | |
4HIV
 
 | | Structure of actinomycin D d(ATGCGGCAT) complex | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*AP*T)-3') | | Authors: | Lo, Y.S, Tseng, W.H, Hou, M.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats.
Nucleic Acids Res., 41, 2013
|
|
