3EIK
 
 | |
3G9A
 
 | | Green fluorescent protein bound to minimizer nanobody | | Descriptor: | Green fluorescent protein, Minimizer | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-02-13 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K1K
 
 | | Green fluorescent protein bound to enhancer nanobody | | Descriptor: | Enhancer, Green Fluorescent Protein | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-09-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
1XEX
 
 | |
2B2N
 
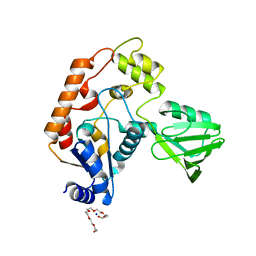 | | Structure of transcription-repair coupling factor | | Descriptor: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, SODIUM ION, SULFATE ION, ... | | Authors: | Assenmacher, N, Wenig, K, Lammens, A, Hopfner, K.-P. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Transcription-coupled Repair: the N Terminus of Mfd Resembles UvrB with Degenerate ATPase Motifs
J.Mol.Biol., 355, 2006
|
|
1XEW
 
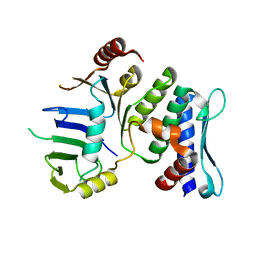 | |
1XBS
 
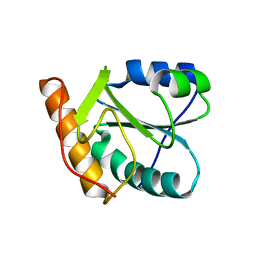 | | Crystal structure of human dim2: a dim1-like protein | | Descriptor: | Dim1-like protein | | Authors: | Simeoni, F, Arvai, A, Hopfner, K.-P, Bello, P, Gondeau, C, Heitz, F, Tainer, J, Divita, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Characterization and Crystal Structure of a Dim1 Family Associated Protein: Dim2
Biochemistry, 44, 2005
|
|
4Y42
 
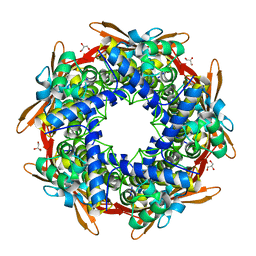 | | Cyanase (CynS) from Serratia proteamaculans | | Descriptor: | Cyanate hydratase, GLYCEROL | | Authors: | Butryn, A, Hopfner, K.-P. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Serendipitous crystallization and structure determination of cyanase (CynS) from Serratia proteamaculans
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3L51
 
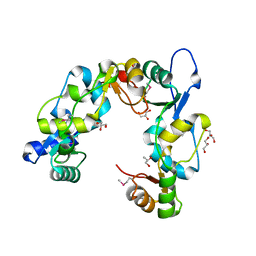 | | Crystal Structure of the Mouse Condensin Hinge Domain | | Descriptor: | GLYCEROL, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Griese, J.J, Hopfner, K.-P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins
Nucleic Acids Res., 38, 2010
|
|
3M7N
 
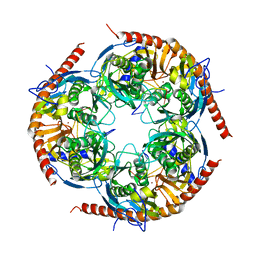 | |
3M85
 
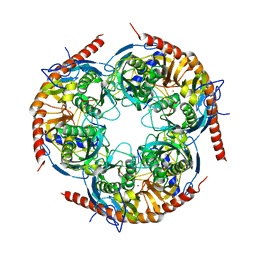 | |
1II8
 
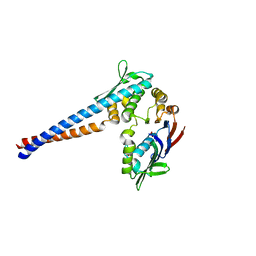 | | Crystal structure of the P. furiosus Rad50 ATPase domain | | Descriptor: | PHOSPHATE ION, Rad50 ABC-ATPase | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1II7
 
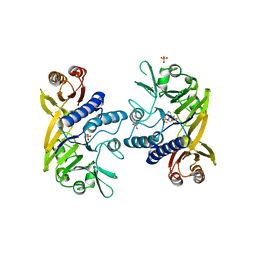 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1RFN
 
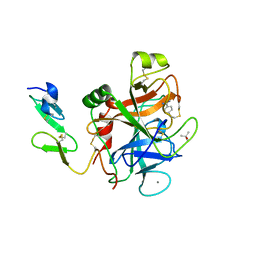 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
1TGO
 
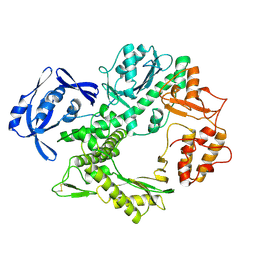 | | THERMOSTABLE B TYPE DNA POLYMERASE FROM THERMOCOCCUS GORGONARIUS | | Descriptor: | PROTEIN (THERMOSTABLE B DNA POLYMERASE) | | Authors: | Hopfner, K.-P, Eichinger, A, Engh, R.A, Laue, F, Ankenbauer, W, Huber, R, Angerer, B. | | Deposit date: | 1999-02-23 | | Release date: | 1999-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4WVY
 
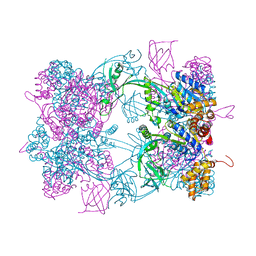 | |
3PP4
 
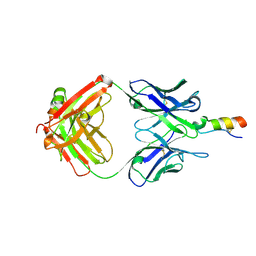 | |
3PP3
 
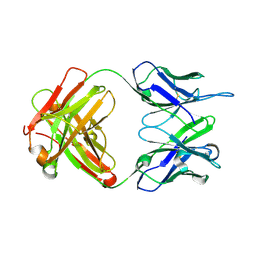 | |
4WK1
 
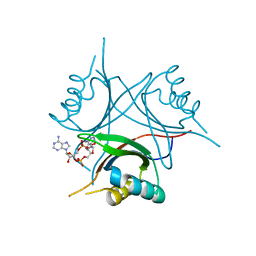 | | Crystal structure of Staphylococcus aureus PstA in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CALCIUM ION, PstA | | Authors: | Mueller, M, Hopfner, K.-P, Witte, G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | c-di-AMP recognition by Staphylococcus aureus PstA.
Febs Lett., 589, 2015
|
|
4WK3
 
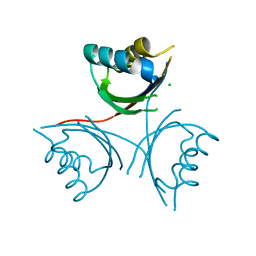 | |
4KB6
 
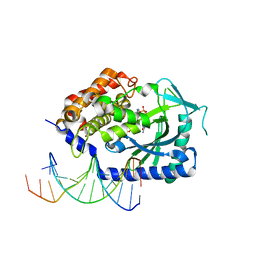 | |
5N6I
 
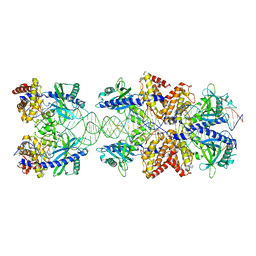 | | Crystal structure of mouse cGAS in complex with 39 bp DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA (36-MER), DNA (37-MER), ... | | Authors: | Andreeva, L, Kostrewa, D, Hopfner, K.-P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders.
Nature, 549, 2017
|
|
6G7E
 
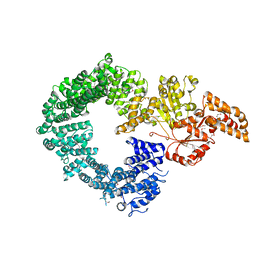 | |
3TBK
 
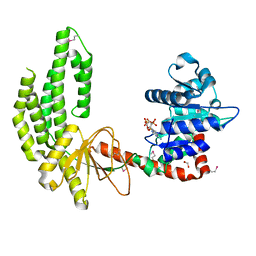 | | Mouse RIG-I ATPase Domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RIG-I Helicase Domain | | Authors: | Civril, F, Bennett, M.D, Hopfner, K.-P. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Embo Rep., 12, 2011
|
|
3TJ1
 
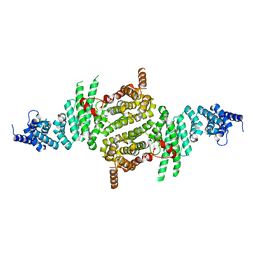 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
