1AEL
 
 | |
1URE
 
 | |
1A57
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A HELIX-LESS VARIANT OF INTESTINAL FATTY ACID BINDING PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN | | Authors: | Steele, R.A, Emmert, D.A, Kao, J, Hodsdon, M.E, Frieden, C, Cistola, D.P. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein.
Protein Sci., 7, 1998
|
|
2AL3
 
 | | Solution structure and backbone dynamics of an N-terminal ubiquitin-like domain in the GLUT4-tethering protein, TUG | | Descriptor: | TUG long isoform | | Authors: | Tettamanzi, M.C, Yu, C, Bogan, J.S, Hodsdon, M.E. | | Deposit date: | 2005-08-04 | | Release date: | 2006-03-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of an N-terminal ubiquitin-like domain in the GLUT4-regulating protein, TUG.
Protein Sci., 15, 2006
|
|
2AGA
 
 | | De-ubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Mao, Y, Senic-Matuglia, F, Di Fiore, P, Polo, S, Hodsdon, M.E, De Camilli, P. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1PJZ
 
 | |
2KIG
 
 | |
2KIE
 
 | | A PH domain within OCRL bridges clathrin mediated membrane trafficking to phosphoinositide metabolis | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1 | | Authors: | Mao, Y, Balkin, D.M, Zoncu, R, Erdmann, K, Tomasini, L, Hu, F, Jin, M.M, Hodsdon, M.E, De Camilli, P. | | Deposit date: | 2009-05-03 | | Release date: | 2009-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism
Embo J., 28, 2009
|
|
2KQ6
 
 | | The structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity | | Descriptor: | Polycystin-2 | | Authors: | Petri, E.T, Celic, A, Kennedy, S.D, Ehrlich, B.E, Boggon, T.J, Hodsdon, M.E. | | Deposit date: | 2009-10-28 | | Release date: | 2010-05-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2MHH
 
 | | Solution structure of a EF-hand domain from sea urchin polycystin-2 | | Descriptor: | Polycystic kidney disease protein 2 | | Authors: | Kuo, I.Y, Keeler, C, Corbin, R, Celic, A, Petri, E.T, Hodsdon, M.E, Ehrlich, B.E. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The number and location of EF hand motifs dictates the calcium dependence of polycystin-2 function.
Faseb J., 28, 2014
|
|
3MZG
 
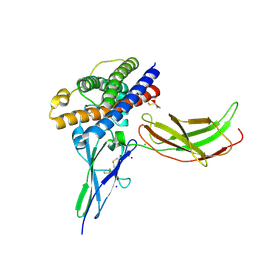 | | Crystal structure of a human prolactin receptor antagonist in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3N06
 
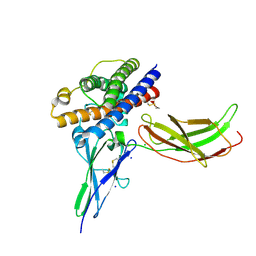 | | A mutant human Prolactin receptor antagonist H27A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3N0P
 
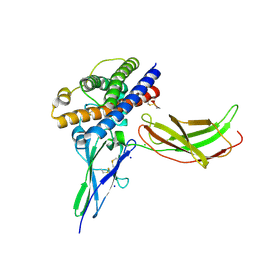 | | A mutant human Prolactin receptor antagonist H30A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-05-14 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3NCE
 
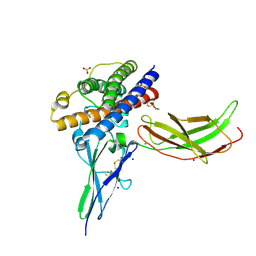 | | A mutant human Prolactin receptor antagonist H27A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3NCF
 
 | | A mutant human Prolactin receptor antagonist H30A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3NCB
 
 | | A mutant human Prolactin receptor antagonist H180A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3NCC
 
 | | A human Prolactin receptor antagonist in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
3IJJ
 
 | | Ternary Complex of Macrophage Migration Inhibitory Factor (MIF) Bound Both to 4-hydroxyphenylpyruvate and to the Allosteric Inhibitor AV1013 (R-stereoisomer) | | Descriptor: | (2E)-2-hydroxy-3-(4-hydroxyphenyl)prop-2-enoic acid, (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ... | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IJG
 
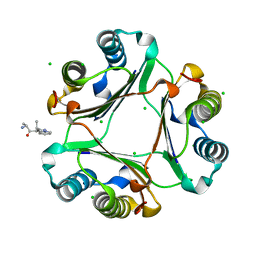 | | Macrophage Migration Inhibitory Factor (MIF) Bound to the (R)-Stereoisomer of AV1013 | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2NWG
 
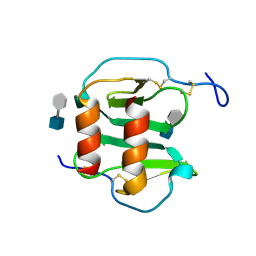 | | Structure of CXCL12:heparin disaccharide complex | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Stromal cell-derived factor 1 | | Authors: | Murphy, J.W, Cho, Y, Lolis, E. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and Functional Basis of CXCL12 (Stromal Cell-derived Factor-1{alpha}) Binding to Heparin
J.Biol.Chem., 282, 2007
|
|
3N52
 
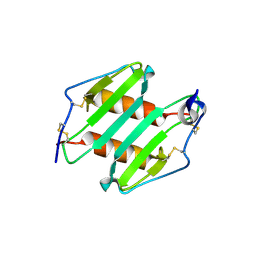 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
