4BR1
 
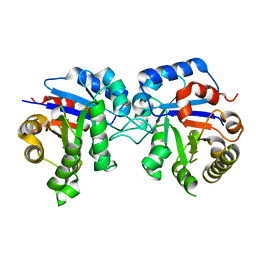 | | Protease-induced heterodimer of human triosephosphate isomerase. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Hernandez-Alcantara, G, Mendoza-Hernandez, G, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Garcia-Torres, I, Torres-Arroyo, A, Gomez-Manzo, S, Marcial-Quino, J, Oria-Hernandez, J, Lopez-Velazquez, G, Reyes-Vivas, H. | | Deposit date: | 2013-06-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Proteolysis in Human Triosephosphate Isomerase
To be Published
|
|
1YRR
 
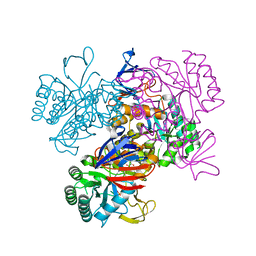 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
4OM9
 
 | | X-Ray Crystal Structure of the passenger domain of Plasmid encoded toxin, an Autrotansporter Enterotoxin from enteroaggregative Escherichia coli (EAEC) | | Descriptor: | Serine protease pet | | Authors: | Meza-Aguilar, J.D, Fromme, P, Torres-Larios, A, Mendoza-Hernandez, G, Hernandez-Chinas, U, Arreguin-Espinosa de Los Monteros, R.A, Eslava-Campos, C.A, Fromme, R. | | Deposit date: | 2014-01-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the passenger domain of plasmid encoded toxin(Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC).
Biochem.Biophys.Res.Commun., 445, 2014
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
3DNJ
 
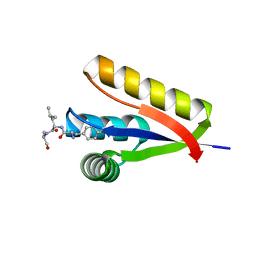 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
2PVX
 
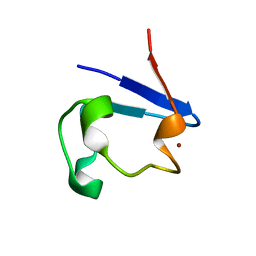 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | Rubredoxin, ZINC ION | | Authors: | Wang, L, LeMaster, D.M, Hernandez, G, Li, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin
Bmc Struct.Biol., 7, 2007
|
|
2PVE
 
 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Rubredoxin, ... | | Authors: | LeMaster, D.M, Anderson, J.S, Wang, L, Guo, Y, Li, H, Hernandez, G. | | Deposit date: | 2007-05-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.79 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin.
Bmc Struct.Biol., 7, 2007
|
|
3G1B
 
 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | Descriptor: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G19
 
 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G3P
 
 | |
3GQ0
 
 | |
3GW1
 
 | | The structure of the Caulobacter crescentus CLPs protease adaptor protein in complex with FGG tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, FGG peptide, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GQ1
 
 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3O1F
 
 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | Descriptor: | ATP-dependent Clp protease adapter protein clpS | | Authors: | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
4N19
 
 | | Structural basis of conformational transitions in the active site and 80 s loop in the FK506 binding protein FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Mustafi, S.M, Brecher, M.B, Zhang, J, Li, H.M, Lemaster, D.M, Hernandez, G. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of conformational transitions in the active site and 80's loop in the FK506-binding protein FKBP12.
Biochem.J., 458, 2014
|
|
4IPX
 
 | | Analyzing the visible conformational substates of the FK506 binding protein FKBP12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysing the visible conformational substates of the FK506-binding protein FKBP12.
Biochem.J., 453, 2013
|
|
4IQ2
 
 | | P21 crystal form of FKBP12.6 | | Descriptor: | MALONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IQC
 
 | | P3121 crystal form of FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6VSI
 
 | | Crystal structure of FKBP12 of Candida auris | | Descriptor: | Peptidylprolyl isomerase, SULFATE ION | | Authors: | Li, Z, Li, H, Hernandez, G, LeMaster, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and transient dimerization for the FKBP12 protein from the pathogenic fungus Candida auris.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
3PF3
 
 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | Deposit date: | 2010-10-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
2DP3
 
 | |
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
