4YN5
 
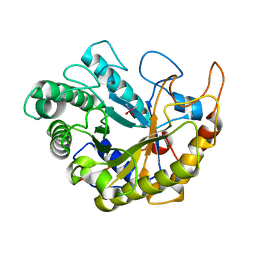 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | Descriptor: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | Authors: | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
1WKY
 
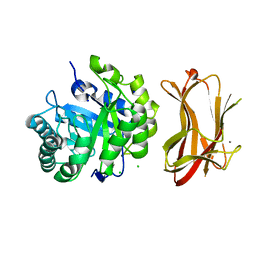 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3VSS
 
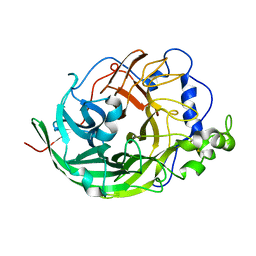 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSR
 
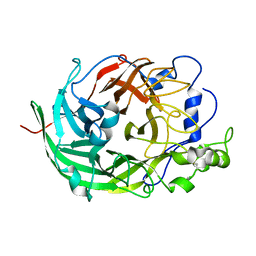 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3WZ1
 
 | | Catalytic domain of beta-agarase from Microbulbifer thermotolerans JAMB-A94 | | Descriptor: | Agarase, GLYCEROL, SODIUM ION | | Authors: | Takagi, E, Hatada, Y, Akita, M, Ohta, Y, Yokoi, G, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the catalytic domain of a GH16 beta-agarase from a deep-sea bacterium, Microbulbifer thermotolerans JAMB-A94
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WPU
 
 | | Full-length beta-fructofuranosidase from Microbacterium saccharophilum K-1 | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPV
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/F447V/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPY
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447V/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPZ
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447P/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
2ZE0
 
 | | Alpha-glucosidase GSJ | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Shirai, T, Hung, V.S, Morinaka, K, Kobayashi, T, Ito, S. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GH13 alpha-glucosidase GSJ from one of the deepest sea bacteria
Proteins, 73, 2008
|
|
