1RY1
 
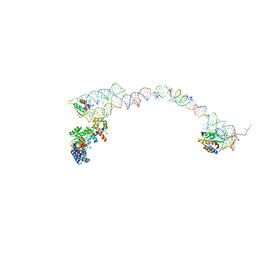 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | Descriptor: | SRP Alu domain, SRP RNA, SRP S domain, ... | | Authors: | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
1YSH
 
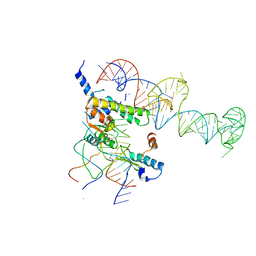 | | Localization and dynamic behavior of ribosomal protein L30e | | Descriptor: | 40S RIBOSOMAL PROTEIN S13, RNA (101-MER), RNA (28-MER), ... | | Authors: | Halic, M, Becker, T, Frank, J, Spahn, C.M, Beckmann, R. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Localization and dynamic behavior of ribosomal protein L30e
Nat.Struct.Mol.Biol., 12, 2005
|
|
2GO5
 
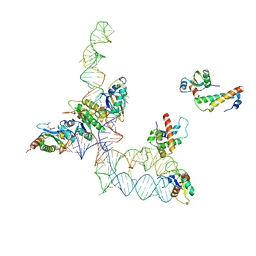 | | Structure of signal recognition particle receptor (SR) in complex with signal recognition particle (SRP) and ribosome nascent chain complex | | Descriptor: | SRP RNA, Signal recognition particle 19 kDa protein (SRP19), Signal recognition particle 54 kDa protein (SRP54), ... | | Authors: | Halic, M, Gartmann, M, Schlenker, O, Mielke, T, Pool, M.R, Sinning, I, Beckmann, R. | | Deposit date: | 2006-04-12 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Signal Recognition Particle Receptor Exposes the Ribosomal Translocon Binding Site
Science, 312, 2006
|
|
2J28
 
 | | MODEL OF E. COLI SRP BOUND TO 70S RNCS | | Descriptor: | 23S RIBOSOMAL RNA, 4.5S SIGNAL RECOGNITION PARTICLE RNA, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-08 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Following the Signal Sequence from Ribosomal Tunnel Exit to Signal Recognition Particle
Nature, 444, 2006
|
|
2J37
 
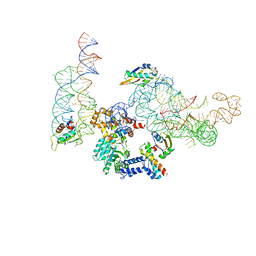 | | MODEL OF MAMMALIAN SRP BOUND TO 80S RNCS | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, RIBOSOMAL PROTEIN L31, RIBOSOMAL PROTEIN L35, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-18 | | Release date: | 2006-11-08 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Following the signal sequence from ribosomal tunnel exit to signal recognition particle.
Nature, 444, 2006
|
|
6NZO
 
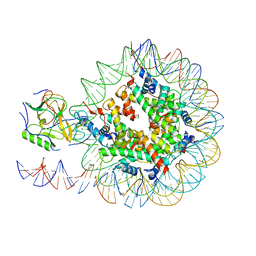 | | Set2 bound to nucleosome | | Descriptor: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | Authors: | Halic, M, Bilokapic, S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6PX1
 
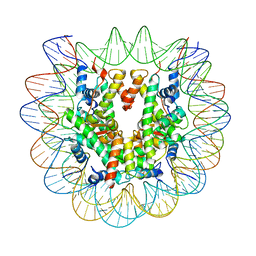 | | Set2 bound to nucleosome | | Descriptor: | DNA (149-MER), Histone H2B 1.1, Histone H3, ... | | Authors: | Halic, M, Bilokapic, S. | | Deposit date: | 2019-07-24 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6PX3
 
 | | Set2 bound to nucleosome | | Descriptor: | DNA (145-MER), Histone H2B 1.1, Histone H3, ... | | Authors: | Halic, M, Bilokapic, S. | | Deposit date: | 2019-07-24 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Nucleosome and ubiquitin position Set2 to methylate H3K36.
Nat Commun, 10, 2019
|
|
6WZ9
 
 | |
6X0L
 
 | |
6X0M
 
 | |
6WZ5
 
 | |
6X0N
 
 | |
6N88
 
 | |
6N89
 
 | |
8EUI
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESR
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETJ
 
 | | Fkbp39 associated 60S nascent ribosome State 2 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUG
 
 | | Ytm1 associated nascent 60S ribosome State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETC
 
 | | Fkbp39 associated nascent 60S ribosome State 4 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-16 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
