1FHB
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE CYANIDE ADDUCT OF A MET80ALA VARIANT OF SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C. IDENTIFICATION OF LIGAND-RESIDUE INTERACTIONS IN THE DISTAL HEME CAVITY | | Descriptor: | CYANIDE ION, FERRICYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1995-06-16 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure of the Cyanide Adduct of a met80Ala Variant of Saccharomyces Cerevisiae Iso-1-Cytochrome C. Identification of Ligand-Residue Interactions in the Distal Heme Cavity
Biochemistry, 34, 1995
|
|
1AKK
 
 | | SOLUTION STRUCTURE OF OXIDIZED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Gray, H.B, Luchinat, C, Reddig, T, Rosato, A, Turano, P. | | Deposit date: | 1997-05-22 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized horse heart cytochrome c.
Biochemistry, 36, 1997
|
|
3NPL
 
 | | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Ener, M, Lee, Y.-T, Goodin, D.B, Winkler, J.R, Gray, H.B, Cheruzel, L. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant
To be Published
|
|
3NP4
 
 | | C112D/M121E Pseudomonas aeruginosa Azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2010-06-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Outer-Sphere Effects on Reduction Potentials of Copper Sites in Proteins: The Curious Case of High Potential Type 2 C112D/M121E Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 132, 2010
|
|
3NP3
 
 | | C112D/M121E Pseudomonas Aeruginosa Azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2010-06-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Outer-Sphere Effects on Reduction Potentials of Copper Sites in Proteins: The Curious Case of High Potential Type 2 C112D/M121E Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 132, 2010
|
|
3OQR
 
 | | C112D/M121E Azurin, pH 10.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Outer-Sphere Effects on Reduction Potentials of Copper Sites in Proteins: The Curious Case of High Potential Type 2 C112D/M121E Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 132, 2010
|
|
3U25
 
 | | Crystal structure of P. aeruginoas azurin containing a Tyr-His hydrogen bonded pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Warren, J.J, Winkler, J.R, Gray, H.B. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Redox properties of tyrosine and related molecules.
Febs Lett., 586, 2012
|
|
3VNW
 
 | | Crystal structure of cytochrome c552 from Thermus thermophilus at pH 5.44 | | Descriptor: | Cytochrome c-552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yamada, S, Bouley-Ford, N.D, Keller, G.E, Winkler, J.R, Gray, H.B. | | Deposit date: | 2012-01-18 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Snapshots of a protein folding intermediate
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3FQY
 
 | | Azurin C112D | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Type Zero Copper Proteins.
Nat Chem, 1, 2009
|
|
4K9J
 
 | | Structure of Re(CO)3(4,7-dimethyl-phen)(Thr126His)(Lys122Trp)(His83Glu)(Trp48Phe)(Tyr72Phe)(Tyr108Phe)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Williamson, H.R, Blanco-Rodriguez, A.M, Sokolova, L, Nikolovski, P, Kaiser, J.T, Towrie, M, Clark, I.P, Vlcek Jr, A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2013-04-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan-accelerated electron flow across a protein-protein interface.
J.Am.Chem.Soc., 135, 2013
|
|
2BC5
 
 | | Crystal structure of E. coli cytochrome b562 with engineered c-type heme linkages | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562 | | Authors: | Faraone-Mennella, J, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stability and Folding Kinetics of Structurally Characterized Cytochrome c-b(562).
Biochemistry, 45, 2006
|
|
2BT3
 
 | | AGAO in complex with Ruthenium-C4-wire at 1.73 angstroms | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Reversible Inhibition of Copper Amine Oxidase Activity by Channel-Blocking Ruthenium(II) and Rhenium(I) Molecular Wires.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2CFD
 
 | | AGAO in complex with wc4l3 (Ru-wire inhibitor, 4-carbon linker, lambda enantiomer, data set 3) | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-20 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CFL
 
 | | AGAO in complex with wc6b (Ru-wire inhibitor, 6-carbon linker, data set b) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-22 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CG1
 
 | | AGAO in complex with wc11b (Ru-wire inhibitor, 11-carbon linker, data set b) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-27 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CFW
 
 | | AGAO in complex with wc7a (Ru-wire inhibitor, 7-carbon linker, data set a) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CFG
 
 | | AGAO in complex with wc4d3 (Ru-wire inhibitor, 4-carbon linker, delta enantiomer, data set 3) | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-21 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CG0
 
 | | AGAO in complex with wc9a (Ru-wire inhibitor, 9-carbon linker, data set a) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-27 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
2CFK
 
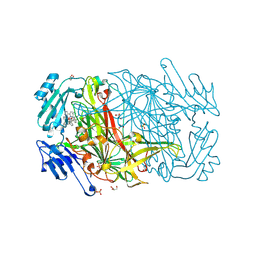 | | AGAO in complex with wc5 (Ru-wire inhibitor, 5-carbon linker) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-21 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enantiomer-specific binding of ruthenium(II) molecular wires by the amine oxidase of Arthrobacter globiformis.
J. Am. Chem. Soc., 130, 2008
|
|
4HHG
 
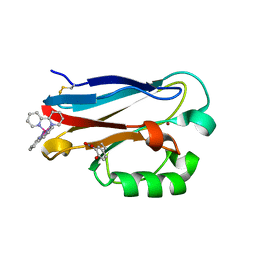 | | Crystal structure of the Pseudomonas aeruginosa azurin, RuH107NO YOH109 | | Descriptor: | Azurin, COPPER (II) ION, DELTA-BIS(2,2'-BIPYRIDINE)IMIDAZOLE RUTHENIUM (II) | | Authors: | Herrera, N, Warren, J.J, Gray, H.B. | | Deposit date: | 2012-10-09 | | Release date: | 2012-11-21 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Electron Flow through Nitrotyrosinate in Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 135, 2013
|
|
4HIP
 
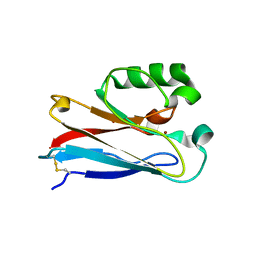 | |
4HHW
 
 | |
1MQV
 
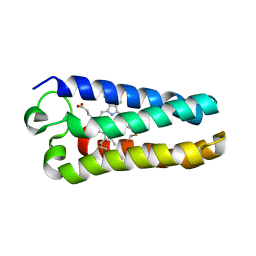 | | Crystal Structure of the Q1A/F32W/W72F mutant of Rhodopseudomonas palustris cytochrome c' (prime) expressed in E. coli | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lee, J.C, Engman, K.C, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Features of Cytochrome c' Folding Intermediates Revealed by Fluorescence Energy-Transfer Kinetics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1QMQ
 
 | | Optical detection of cytochrome P450 by sensitizer-linked substrates | | Descriptor: | ACETATE ION, CYTOCHROME P450, DELTA-BIS(2,2'-BIPYRIDINE)-(5-METHYL-2-2'-BIPYRIDINE)-C9-ADAMANTANE RUTHENIUM (II), ... | | Authors: | Crane, B.R, Dmochowski, I.J, Gray, H.B. | | Deposit date: | 1999-10-05 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Optical Detection of Cytochrome P450 by Sensitizer-Linked Substrates
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RF9
 
 | | Crystal structure of cytochrome P450-cam with a fluorescent probe D-4-AD (Adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-BUTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational States of Cytochrome P450cam Revealed by Trapping of synthetic Molecular Wires
J.Mol.Biol., 344, 2004
|
|
