7QB3
 
 | |
1FEQ
 
 | | NMR SOLUTION STRUCTURE OF THE ANTICODON OF TRNA(LYS3) WITH T6A MODIFICATION AT POSITION 37 | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*CP*UP*UP*UP*UP*(T6A)P*AP*UP*CP*UP*GP*C)-3' | | Authors: | Stuart, J.W, Gdaniec, Z, Guenther, R.H, Marszalek, M, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2000-07-21 | | Release date: | 2000-11-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A.
Biochemistry, 39, 2000
|
|
1XV6
 
 | |
8OR8
 
 | |
6GE1
 
 | |
6I1W
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Gp(PSU)pC-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*GP*AP*CP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*GP*(PSU)P*CP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
6I1V
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Cp(PSU)pG-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*CP*AP*GP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*CP*(PSU)P*GP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
2JXS
 
 | |
2K41
 
 | | NMR structure of uridine bulged RNA duplex | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*UP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2K3Z
 
 | | NMR structure of adenosine bulged RNA duplex with C:G-A triple | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*AP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2JXQ
 
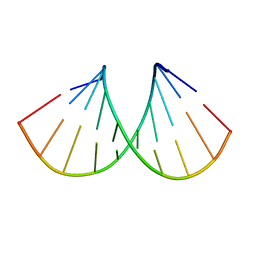 | | NMR structure of RNA duplex | | Descriptor: | RNA (5'-R(*CP*GP*CP*UP*CP*UP*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Popenda, L, Adamiak, R.W, Gdaniec, Z. | | Deposit date: | 2007-11-28 | | Release date: | 2008-05-20 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Bulged adenosine influence on the RNA duplex conformation in solution.
Biochemistry, 47, 2008
|
|
2MCJ
 
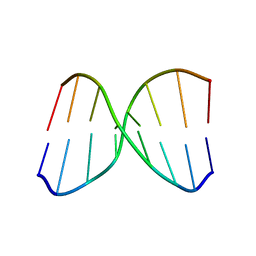 | | NMR structure of spermine modified DNA duplex | | Descriptor: | DNA_(5'-D(*CP*AP*GP*(N4S)P*CP*GP*AP*C)-3'), DNA_(5'-D(*GP*TP*CP*GP*GP*CP*TP*G)-3') | | Authors: | Brzezinska, J, Gdaniec, Z, Popenda, L, Markiewicz, W.T. | | Deposit date: | 2013-08-20 | | Release date: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Polyaminooligonucleotide: NMR structure of duplex DNA containing a nucleoside with spermine residue, N-[4,9,13-triazatridecan-1-yl]-2'-deoxycytidine.
Biochim.Biophys.Acta, 1840, 2014
|
|
2MCI
 
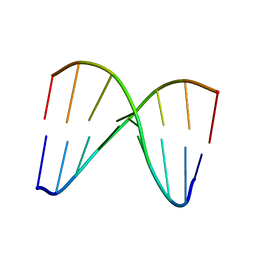 | | NMR structure of DNA duplex | | Descriptor: | DNA_(5'-D(*CP*AP*GP*CP*CP*GP*AP*C)-3'), DNA_(5'-D(*GP*TP*CP*GP*GP*CP*TP*G)-3') | | Authors: | Brzezinska, J, Gdaniec, Z, Popenda, L, Markiewicz, W.T. | | Deposit date: | 2013-08-20 | | Release date: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Polyaminooligonucleotide: NMR structure of duplex DNA containing a nucleoside with spermine residue, N-[4,9,13-triazatridecan-1-yl]-2'-deoxycytidine.
Biochim.Biophys.Acta, 1840, 2014
|
|
6F3P
 
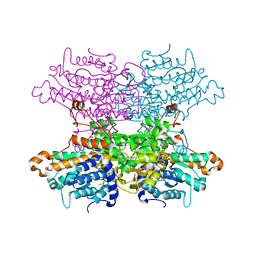 | |
6F3Q
 
 | |
6F3O
 
 | |
6F3N
 
 | |
6F3M
 
 | |
