181D
 
 | |
180D
 
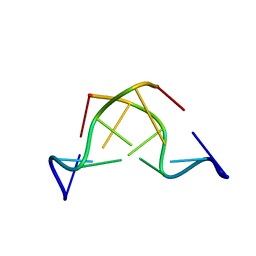 | |
1QQU
 
 | |
1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4OAQ
 
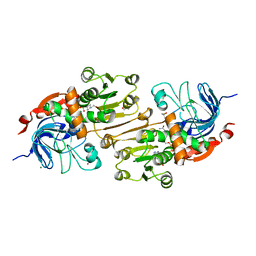 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
4OKL
 
 | |
4ZKK
 
 | | The novel double-fold structure of d(GCATGCATGC) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*CP*AP*TP*GP*CP*AP*TP*GP*C)-3') | | Authors: | Thirugnanasambandam, A, Karthik, S, Mandal, P.K, Gautham, N. | | Deposit date: | 2015-04-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The novel double-folded structure of d(GCATGCATGC): a possible model for triplet-repeat sequences
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WV7
 
 | |
5FHJ
 
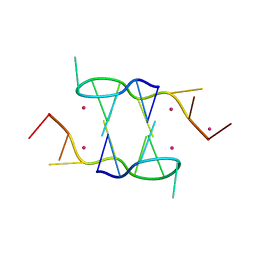 | |
5FHL
 
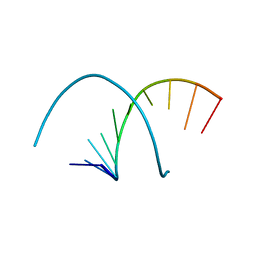 | |
351D
 
 | |
5IYE
 
 | | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 resolution | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*CP*GP*GP*G)-3'), ZINC ION | | Authors: | Karthik, S, Thirugnanasambandam, A, Mandal, P.K, Gautham, N. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 angstrom resolution.
Nucleosides Nucleotides Nucleic Acids, 36, 2017
|
|
5IYG
 
 | |
5IYJ
 
 | |
6A1O
 
 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*G)-3'), ... | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
2HTO
 
 | | Ruthenium hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), RUTHENIUM (III) HEXAAMINE ION | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2HTT
 
 | | Ruthenium Hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DTP*DG)-3'), ... | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
6A1Q
 
 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*G)-3') | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
1XA2
 
 | |
1XAM
 
 | | Cobalt hexammine induced tautameric shift in Z-DNA: structure of d(CGCGCA).d(TGCGCG) in two crystal forms. | | Descriptor: | CGCGCA, COBALT HEXAMMINE(III), TG, ... | | Authors: | Thiyagarajan, S, Rajan, S.S, Gautham, N. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cobalt hexammine induced tautomeric shift in Z-DNA: the structure of d(CGCGCA)*d(TGCGCG) in two crystal forms.
Nucleic Acids Res., 32, 2004
|
|
240D
 
 | |
239D
 
 | |
3HQE
 
 | |
3HS1
 
 | |
3IXN
 
 | |
