6IWB
 
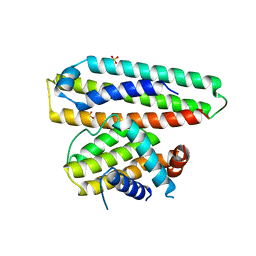 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
7XYQ
 
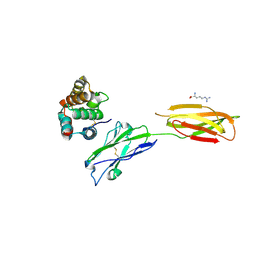 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | Descriptor: | ARGININE, CD274 molecule, DBL1_03 | | Authors: | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XAD
 
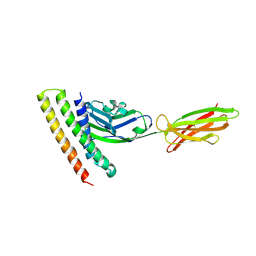 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7AYE
 
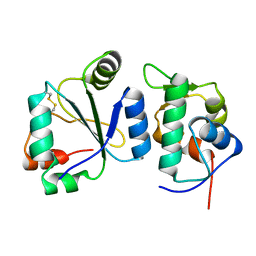 | | Crystal structure of the computationally designed chemically disruptable heterodimer LD6-MDM2 | | Descriptor: | Isoform 11 of E3 ubiquitin-protein ligase Mdm2, Thiol:disulfide interchange protein DsbD | | Authors: | Yang, C, Lau, K, Pojer, F, Correia, B.E. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A rational blueprint for the design of chemically-controlled protein switches.
Nat Commun, 12, 2021
|
|
7T7S
 
 | | R-27 in Complex with S. aureus DHFR and tricyclic-NADPH (tNADPH) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
7T7Q
 
 | | R-27 In Complex with S. aureus DHFR and alpha-NADPH - Remediated for comparison with tNADPH | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
4Q6A
 
 | |
4Q67
 
 | |
