1AT9
 
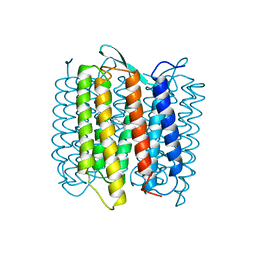 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2011-07-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
4UX1
 
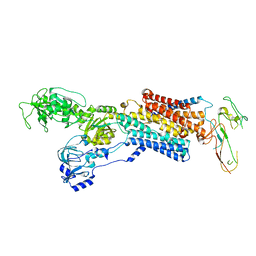 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4BGN
 
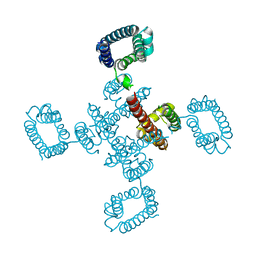 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
1OED
 
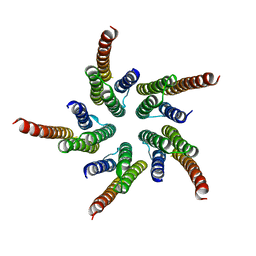 | | STRUCTURE OF ACETYLCHOLINE RECEPTOR PORE FROM ELECTRON IMAGES | | Descriptor: | Acetylcholine receptor beta subunit, Acetylcholine receptor delta subunit, Acetylcholine receptor gamma subunit, ... | | Authors: | Miyazawa, A, Fujiyoshi, Y, Unwin, N. | | Deposit date: | 2003-03-24 | | Release date: | 2003-06-26 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and gating mechanism of the acetylcholine receptor pore.
Nature, 423, 2003
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
1L9H
 
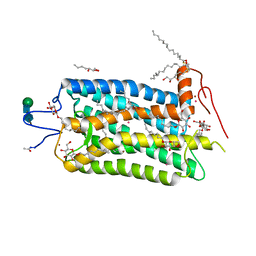 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1I2H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSD-ZIP45(HOMER1C/VESL-1L)CONSERVED HOMER 1 DOMAIN | | Descriptor: | PSD-ZIP45(HOMER-1C/VESL-1L) | | Authors: | Irie, K, Nakatsu, T, Mitsuoka, K, Fujiyoshi, Y, Kato, H. | | Deposit date: | 2001-02-09 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Homer 1 Family Conserved Region Reveals the Interaction Between the EVH1 Domain and
Own Proline-rich Motif
J.Mol.Biol., 318, 2002
|
|
3IYZ
 
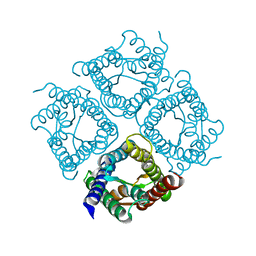 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
1FQY
 
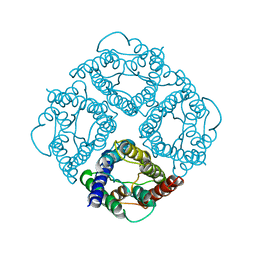 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
2AT9
 
 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
7WSV
 
 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2ZZ9
 
 | | Structure of aquaporin-4 S180D mutant at 2.8 A resolution by electron crystallography | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aquaporin-4 | | Authors: | Tani, K, Mitsuma, T, Hiroaki, Y, Kamegawa, A, Nishikawa, K, Tanimura, Y, Fujiyoshi, Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion.
J.Mol.Biol., 389, 2009
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | Descriptor: | Claudin-19, Heat-labile enterotoxin B chain | | Authors: | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
2B6P
 
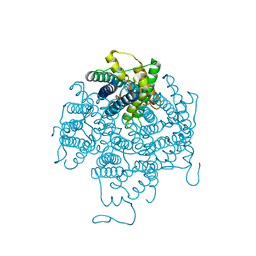 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2B6O
 
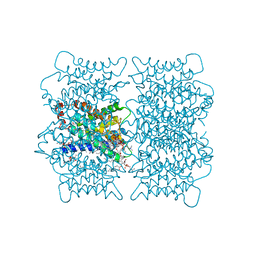 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
3IXZ
 
 | | Pig gastric H+/K+-ATPase complexed with aluminium fluoride | | Descriptor: | Potassium-transporting ATPase alpha, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Tani, K, Nishizawa, T, Fujiyoshi, Y. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.5 Å) | | Cite: | Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle
Embo J., 28, 2009
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
5GLH
 
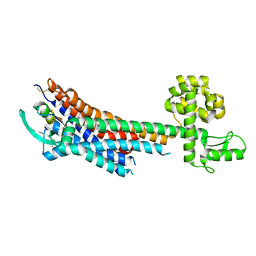 | | Human endothelin receptor type-B in complex with ET-1 | | Descriptor: | Endothelin Receptor Subtype-B, Peptide from Endothelin-1 | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
5H1R
 
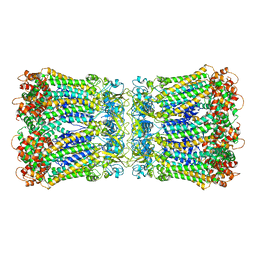 | |
