1M6J
 
 | |
4AJ0
 
 | | Crystallographic structure of an amyloidogenic variant, 3rCW, of the germinal line lambda 3 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, GERMINAL LINE LAMBDA 3 3RCW VARIANT | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
6CG9
 
 | | Crystal structure of Triosephosphate Isomerase from Zea mays (mexican corn) | | Descriptor: | ACETATE ION, GLYCEROL, Triosephosphate isomerase, ... | | Authors: | Romero-Romero, S, Fernandez-Velasco, D.A, Rodriguez-Romero, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and conformational stability of the triosephosphate isomerase from Zea mays. Comparison with the chemical unfolding pathways of other eukaryotic TIMs.
Arch. Biochem. Biophys., 658, 2018
|
|
6YQY
 
 | | Crystal structure of sTIM11noCys, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel sTIM11noCys | | Authors: | Romero-Romero, S, Wiese, G.J, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6YQX
 
 | | Crystal structure of DeNovoTIM13, a de novo designed TIM barrel | | Descriptor: | CHLORIDE ION, GLYCEROL, de novo designed TIM barrel DeNovoTIM13 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6Z2I
 
 | | Crystal structure of DeNovoTIM6, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel DeNovoTIM6 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
4AIX
 
 | | Crystallographic structure of an amyloidogenic variant, 3rC34Y, of the germinal line lambda 3 | | Descriptor: | IG LAMBDA CHAIN V-IV REGION BAU | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AIZ
 
 | | Crystallographic structure of 3mJL2 from the germinal line lambda 3 | | Descriptor: | 1,5:6,10-dianhydro-3,4,7,8-tetradeoxy-2,9-bis-C-(hydroxymethyl)-L-manno-decitol, CITRIC ACID, SULFATE ION, ... | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
6NEE
 
 | |
6XKS
 
 | | Crystal structure of domain A from the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) of Salmonella typhimurium | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Histidine ABC transporter substrate-binding protein HisJ | | Authors: | Romero-Romero, S, Berrocal, T, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and kinetic analysis of the LAO binding protein and its isolated domains reveal non-additivity in stability, folding and function.
Febs J., 2023
|
|
5IR3
 
 | |
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | Descriptor: | CEMP1-p1 | | Authors: | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
5VMR
 
 | |
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
4Y9A
 
 | |
4Y8F
 
 | |
4Y96
 
 | | Crystal structure of Triosephosphate Isomerase from Gemmata obscuriglobus | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
5C9K
 
 | |
6NXX
 
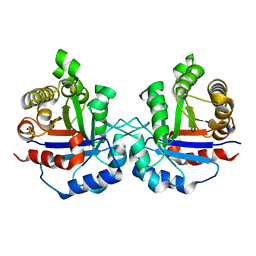 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218K mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Fuentes-Pascacio, A, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXQ
 
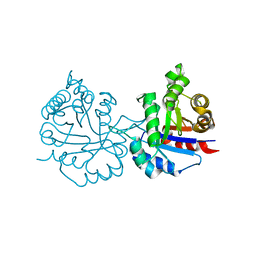 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C13A mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXW
 
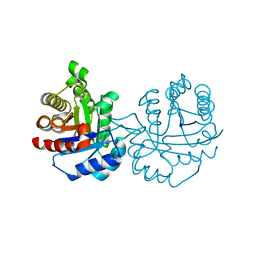 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218S mutant | | Descriptor: | Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXR
 
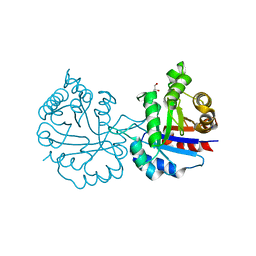 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C13D mutant | | Descriptor: | GLYCEROL, SODIUM ION, Triosephosphate isomerase, ... | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXY
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218D mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXS
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218Y mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
