4ZNM
 
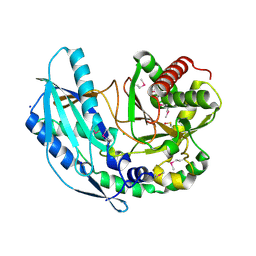 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZXW
 
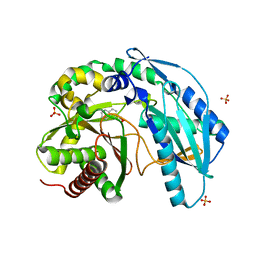 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
5CQF
 
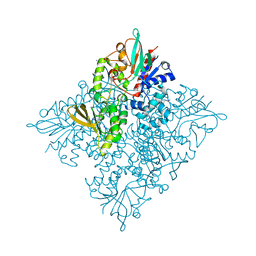 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5BMO
 
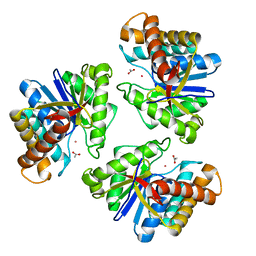 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
5DU2
 
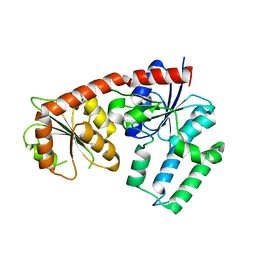 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
5W27
 
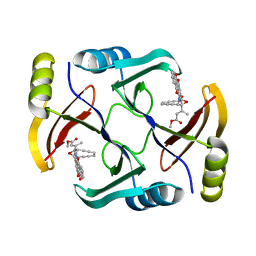 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
6WF4
 
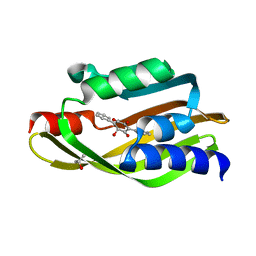 | | Crystal Structure of TerC Co-crystallized with Polyporic Acid | | Descriptor: | (2~5~S)-2~3~,2~5~,2~6~-trihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~(2~5~H)-one, ISOPROPYL ALCOHOL, Terfestatin Biosyntheis Enzyme C | | Authors: | Clinger, J.A, Miller, M.D, Hall, R.E, Zhang, Y, Elshahawi, S.I, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of two cooperative enzymes responsible for the stability
of p-terphenyls.
To be published
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
7N7V
 
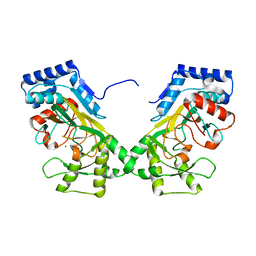 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
5JR3
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
4QA9
 
 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | Descriptor: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | Authors: | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5JXM
 
 | | Crystal Structure of Prenyltransferase PriB Apo Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTH
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | Descriptor: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
