1C6S
 
 | | THE SOLUTION STRUCTURE OF CYTOCHROME C6 FROM THE THERMOPHILIC CYANOBACTERIUM SYNECHOCOCCUS ELONGATUS, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Roesch, P, Beissinger, M, Sticht, H, Sutter, M, Ejchart, A, Haehnel, W. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome c6 from the thermophilic cyanobacterium Synechococcus elongatus.
EMBO J., 17, 1998
|
|
1NKF
 
 | | CALCIUM-BINDING PEPTIDE, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM-BINDING HEXADECAPEPTIDE, LANTHANUM (III) ION | | Authors: | Sticht, H, Ejchart, A. | | Deposit date: | 1998-03-09 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix nucleation by a calcium-binding peptide loop.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DIP
 
 | | THE SOLUTION STRUCTURE OF PORCINE DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE | | Authors: | Roesch, P, Seidel, G, Adermann, K, Schindler, T, Ejchart, A, Jaenicke, R, Forssmann, W.G. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product.
J.Biol.Chem., 272, 1997
|
|
2L0P
 
 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2017-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
2LP3
 
 | | Solution structure of S100A1 Ca2+ | | Descriptor: | CALCIUM ION, Protein S100-A1 | | Authors: | Budzinska, M, Ruszczynska-Bartnik, K, Belczyk-Ciesielska, A, Bierzynski, A, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2014-08-27 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2JPT
 
 | |
2LHL
 
 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
2LLS
 
 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
2LP2
 
 | | Solution structure and dynamics of human S100A1 protein modified at cysteine 85 with homocysteine disulfide bond formation in calcium saturated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, CALCIUM ION, Protein S100-A1 | | Authors: | Nowakowski, M.E, Jaremko, L, Jaremko, M, Zdanowski, K, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2LUX
 
 | |
2M1I
 
 | |
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
1TVS
 
 | |
5LWC
 
 | | NMR solution structure of bacteriocin BacSp222 from Staphylococcus pseudintermedius 222 | | Descriptor: | Bacteriocin BacSp222 | | Authors: | Nowakowski, M.E, Ejchart, A.O, Jaremko, L, Wladyka, B, Mak, P. | | Deposit date: | 2016-09-15 | | Release date: | 2017-10-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Spatial attributes of the four-helix bundle group of bacteriocins - The high-resolution structure of BacSp222 in solution.
Int.J.Biol.Macromol., 107, 2018
|
|
1B6F
 
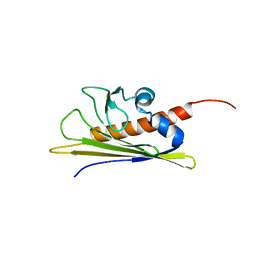 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | PROTEIN (MAJOR POLLEN ALLERGEN BET V 1-A) | | Authors: | Schweimer, K, Sticht, H, Boehm, M, Roesch, P. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Spectroscopy Reveals Common Structural Features of the Birch Pollen Allergen Bet v 1 and the cherry allergen Pru a 1
APPL.MAGN.RESON., 17, 1999
|
|
1HPH
 
 | |
2RQS
 
 | | 3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin) | | Descriptor: | Parvulin-like peptidyl-prolyl isomerase | | Authors: | Zhukov, I, Jaremko, L, Jaremko, M, Mueller, J.W, Bayer, P. | | Deposit date: | 2009-11-17 | | Release date: | 2010-11-24 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the First Archaeal Parvulin Reveal a New Functionally Important Loop in Parvulin-type Prolyl Isomerases
J.Biol.Chem., 286, 2011
|
|
1TIV
 
 | |
2M3W
 
 | |
2V1V
 
 | |
1TAC
 
 | |
1TBC
 
 | |
