5UGQ
 
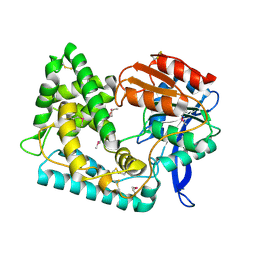 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5UOH
 
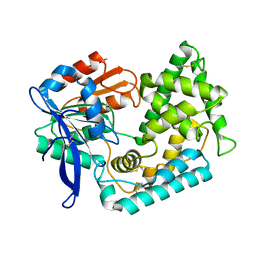 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5UNO
 
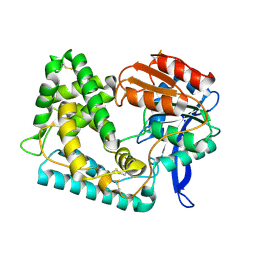 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
1G0V
 
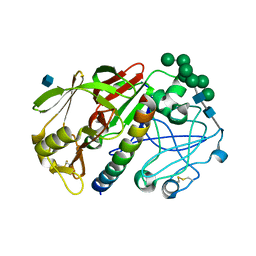 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
5BKM
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine)
To Be Published
|
|
6M9C
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Pseudotyrostatin | | Descriptor: | ACETIC ACID, CALCIUM ION, Pseudotyrostatin, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M9D
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Chymostatin | | Descriptor: | CALCIUM ION, Chymostatin A, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M9F
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Tyrostatin | | Descriptor: | CALCIUM ION, SEDOLISIN, SULFATE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8Y
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIPF | | Descriptor: | AIPF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8W
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIAF | | Descriptor: | AIAF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
1DPJ
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1GTL
 
 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
2R5P
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Indinavir | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, ... | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
2R5Q
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
2R8N
 
 | | Structural Analysis of the Unbound Form of HIV-1 Subtype C Protease | | Descriptor: | GLYCEROL, Pol protein | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-11 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of unbound human immunodeficiency virus 1 subtype C protease: implications of flap dynamics and drug resistance.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2R9B
 
 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
1NLU
 
 | | Pseudomonas sedolisin (serine-carboxyl proteinase) complexed with two molecules of pseudo-iodotyrostatin | | Descriptor: | CALCIUM ION, PSEUDO-IODOTYROSTATIN, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Glodfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Two inhibitor molecules bound in the active site of Pseudomonas sedolisin: a model for the bi-product complex following cleavage of a peptide substrate.
Biochem.Biophys.Res.Commun., 314, 2004
|
|
1SGU
 
 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1SH9
 
 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | POL polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, McKenna, R, Abanje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-25 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
2A8I
 
 | | Crystal Structure of human Taspase1 | | Descriptor: | Threonine aspartase 1 | | Authors: | Khan, J.A, Dunn, B.M, Tong, L. | | Deposit date: | 2005-07-08 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Taspase1, a Crucial Protease Regulating the Function of MLL.
Structure, 13, 2005
|
|
2A8J
 
 | |
