2B0T
 
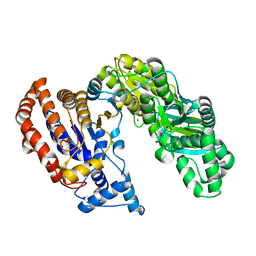 | | Structure of Monomeric NADP Isocitrate dehydrogenase | | Descriptor: | MAGNESIUM ION, NADP Isocitrate dehydrogenase | | Authors: | Imabayashi, F, Aich, S, Prasad, L, Delbaere, L.T. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase.
Proteins, 63, 2006
|
|
1WVQ
 
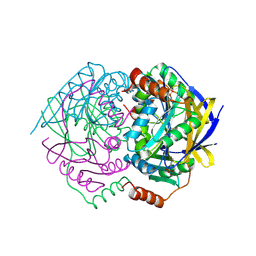 | | Structure of conserved hypothetical protein PAE2307 from Pyrobaculum aerophilum | | Descriptor: | PHOSPHATE ION, hypothetical protein PAE2307 | | Authors: | Lott, J.S, Delbaere, L.T, Banfield, M.J, Sigrell-Simon, J.A, Baker, E.N. | | Deposit date: | 2004-12-24 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of an ancient conserved domain establishes a structural basis for stable histidine phosphorylation and identifies a new family of adenosine-specific kinases.
J.Biol.Chem., 281, 2006
|
|
1YGG
 
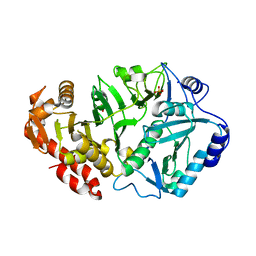 | | Crystal structure of phosphoenolpyruvate carboxykinase from Actinobacillus succinogenes | | Descriptor: | SODIUM ION, SULFATE ION, phosphoenolpyruvate carboxykinase | | Authors: | Leduc, Y.A, Prasad, L, Laivenieks, M, Zeikus, J.G, Delbaere, L.T. | | Deposit date: | 2005-01-04 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YLH
 
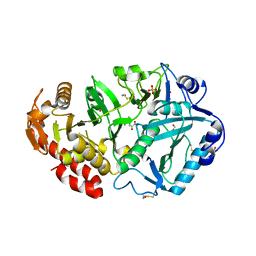 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase from Actinobaccilus succinogenes in Complex with Manganese and Pyruvate | | Descriptor: | (2S,3S)-2,3-DIHYDROXY-4-SULFANYLBUTANE-1-SULFONATE, BETA-MERCAPTOETHANOL, FORMIC ACID, ... | | Authors: | Leduc, Y.A, Prasad, L, Laivenieks, M, Zeikus, J.G, Delbaere, L.T. | | Deposit date: | 2005-01-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YVY
 
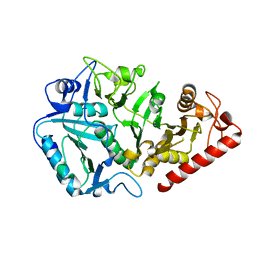 | | Crystal structure of Anaerobiospirillum succiniciproducens phosphoenolpyruvate carboxykinase | | Descriptor: | Phosphoenolpyruvate carboxykinase [ATP] | | Authors: | Cotelesage, J.J, Prasad, L, Zeikus, J.G, Laivenieks, M, Delbaere, L.T. | | Deposit date: | 2005-02-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Anaerobiospirillum succiniciproducens PEP carboxykinase reveals an important active site loop.
Int.J.Biochem.Cell Biol., 37, 2005
|
|
2OLR
 
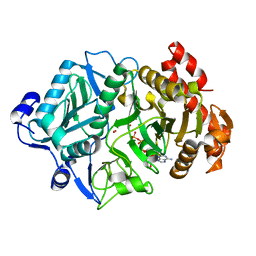 | | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase complexed with carbon dioxide, Mg2+, ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Cotelesage, J.J, Delbaere, L.T, Goldie, H, Puttick, J, Rajabi, B, Novakovski, B. | | Deposit date: | 2007-01-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How does an enzyme recognize CO2?
Int.J.Biochem.Cell Biol., 39, 2007
|
|
2OLQ
 
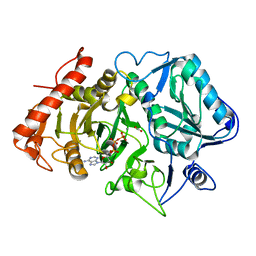 | | How Does an Enzyme Recognize CO2? | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, MAGNESIUM ION, ... | | Authors: | Puttick, J, Goldie, H, Cotelesage, J.J, Rajabi, B, Novakovski, B, Delbaere, L.T. | | Deposit date: | 2007-01-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How does an enzyme recognize CO2?
Int.J.Biochem.Cell Biol., 39, 2007
|
|
1OS1
 
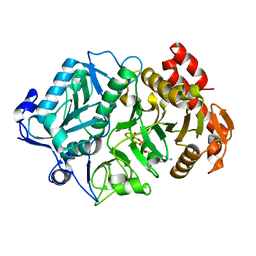 | | Structure of Phosphoenolpyruvate Carboxykinase complexed with ATP,Mg, Ca and pyruvate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sudom, A, Walters, R, Pastushok, L, Goldie, D, Prasad, L, Delbaere, L.T, Goldie, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin.
J.BACTERIOL., 185, 2003
|
|
1P25
 
 | | Crystal structure of nickel(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', NICKEL (II) ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
1P26
 
 | | Crystal structure of zinc(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', ZINC ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
1P24
 
 | | Crystal structure of cobalt(II)-d(GGCGCC)2 | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*GP*CP*GP*CP*C)-3') | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
1LEC
 
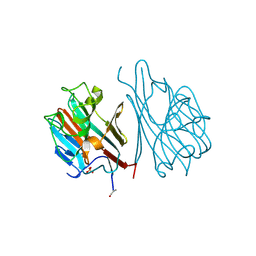 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Delbaere, L, Vandonselaar, M, Quail, J. | | Deposit date: | 1992-12-17 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
1LED
 
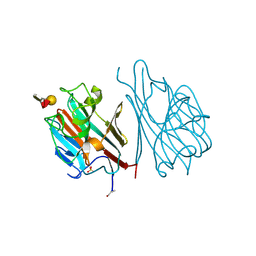 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Delbaere, L, Vandonselaar, M, Quail, J. | | Deposit date: | 1992-12-17 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
1GSL
 
 | |
1SGC
 
 | |
5SGA
 
 | |
4SGA
 
 | |
1AQ2
 
 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tari, L.W, Matte, A, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 1997-08-05 | | Release date: | 1998-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mg(2+)-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions.
Nat.Struct.Biol., 4, 1997
|
|
3SGA
 
 | |
1AYL
 
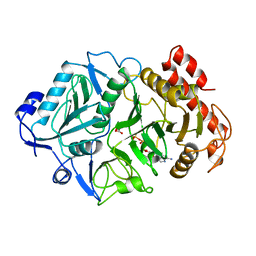 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Tari, L.W, Pugazenthi, U, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 1995-12-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase.
Nat.Struct.Biol., 3, 1996
|
|
3MBC
 
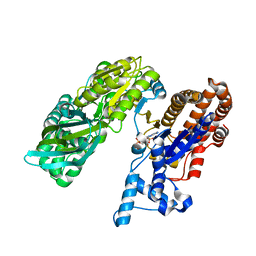 | | Crystal structure of monomeric isocitrate dehydrogenase from Corynebacterium glutamicum in complex with NADP | | Descriptor: | Isocitrate dehydrogenase [NADP], MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sidhu, N.S, Aich, S, Sheldrick, G.M, Delbaere, L.T.J. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly NADP+-specific isocitrate dehydrogenase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ALP
 
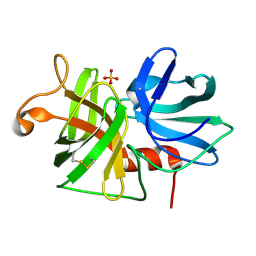 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
1CM2
 
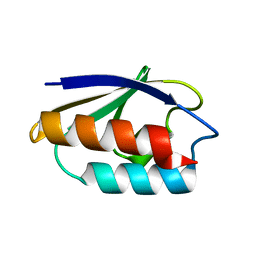 | | STRUCTURE OF HIS15ASP HPR AFTER HYDROLYSIS OF RINGED SPECIES. | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Delbaere, L.T.J, Waygood, E.B. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
1CM3
 
 | | HIS15ASP HPR FROM E. COLI | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Waygood, E.B, Delbaere, L.T.J. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
2JEL
 
 | | JEL42 FAB/HPR COMPLEX | | Descriptor: | HISTIDINE-CONTAINING PROTEIN, JEL42 FAB FRAGMENT, SULFATE ION | | Authors: | Prasad, L, Waygood, E.B, Lee, J.S, Delbaere, L.T.J. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A resolution structure of the jel42 Fab fragment/HPr complex
J.Mol.Biol., 280, 1998
|
|
