6QGS
 
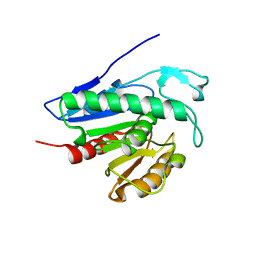 | | Crystal structure of APT1 bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
2HZ8
 
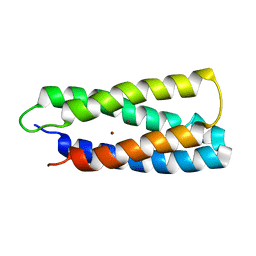 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
6QGQ
 
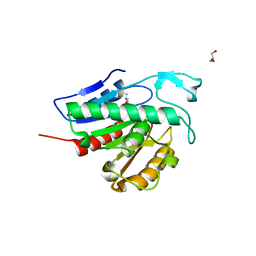 | | Crystal structure of APT1 C2S mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, GLYCEROL, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGN
 
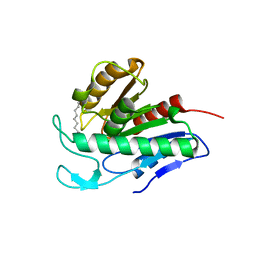 | | Crystal structure of APT1 bound to 2-Bromopalmitate | | Descriptor: | 2-Bromopalmitic acid, Acyl-protein thioesterase 1 | | Authors: | Marcaida, M.J, Audagnotto, M, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGO
 
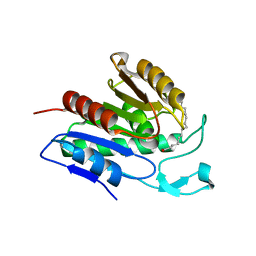 | | Crystal structure of APT1 S119A mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
7OBM
 
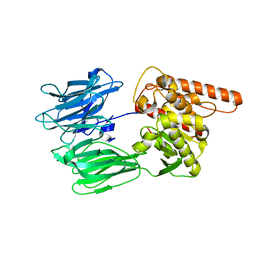 | | Crystal structure of the human Prolyl Endopeptidase-Like protein short form (residues 90-727) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Rosier, K, McDevitt, M.T, Brendan, J.F, Marcaida, M.J, Bingman, C.A, Pagliarini, D.J, Creemers, J.W.M, Smith, R.W, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-10 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function.
Iscience, 24, 2021
|
|
6HTF
 
 | |
6HX0
 
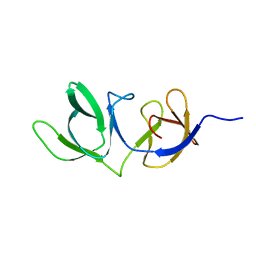 | | Cell wall binding domain of endolysin from Listeria phage A500. | | Descriptor: | L-alanyl-D-glutamate peptidase | | Authors: | Dunne, M, Taylor, N.M.I, Shen, Y, Loessner, M.J, Leiman, P.G. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis for recognition of bacterial cell wall teichoic acid by pseudo-symmetric SH3b-like repeats of a viral peptidoglycan hydrolase
Chem Sci, 2020
|
|
8PBV
 
 | |
4ALZ
 
 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
8CT1
 
 | |
8CT9
 
 | |
7SSP
 
 | |
7SSS
 
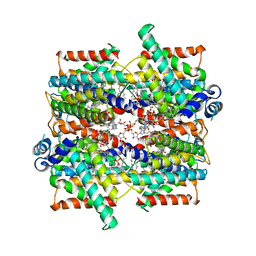 | | Structure of the NADH-bound human COQ7:COQ9 complex by single-particle electron cryo-microscopy | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, 5-demethoxyubiquinone hydroxylase, ... | | Authors: | Aydin, H, Frost, A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and functionality of a multimeric human COQ7:COQ9 complex.
Mol.Cell, 82, 2022
|
|
5I35
 
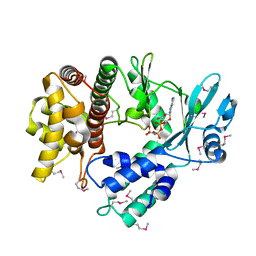 | | Structure of the Human mitochondrial kinase COQ8A R611K with AMPPNP (Cerebellar Ataxia and Ubiquinone Deficiency Through Loss of Unorthodox Kinase Activity) | | Descriptor: | Atypical kinase ADCK3, mitochondrial, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bingman, C.A, Stefely, J.A, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity.
Mol.Cell, 63, 2016
|
|
3C0N
 
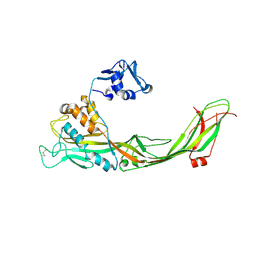 | | Crystal structure of the proaerolysin mutant Y221G at 2.2 A | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0O
 
 | | Crystal structure of the proaerolysin mutant Y221G complexed with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0M
 
 | | Crystal structure of the proaerolysin mutant Y221G | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
6AWL
 
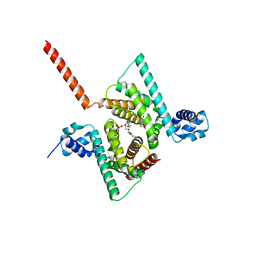 | | Crystal structure of human Coq9 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2017-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol. Cell, 73, 2019
|
|
6DEW
 
 | | Structure of human COQ9 protein with bound isoprene. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2018-05-13 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol.Cell, 73, 2019
|
|
3G4O
 
 | |
3G4N
 
 | |
5JZH
 
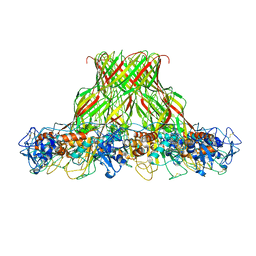 | | Cryo-EM structure of aerolysin prepore | | Descriptor: | Aerolysin | | Authors: | Iacovache, I, Zuber, B. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process.
Nat Commun, 7, 2016
|
|
5JZW
 
 | |
5JZT
 
 | |
