8K4I
 
 | |
8K5Q
 
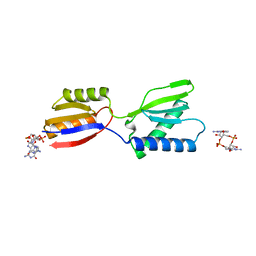 | | Crystal structure of YajQ STM0435 with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), YajQ | | Authors: | Dai, Y, Zhang, M, Wang, W, Li, B. | | Deposit date: | 2023-07-23 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A c-di-GMP binding effector STM0435 modulates flagellar motility and pathogenicity in Salmonella
Virulence, 15, 2024
|
|
8IL0
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 | | Descriptor: | Glycosyltransferase | | Authors: | Dai, Y, Li, P, Qiao, H, Xia, M, Liu, W, Fang, P. | | Deposit date: | 2023-03-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7E8I
 
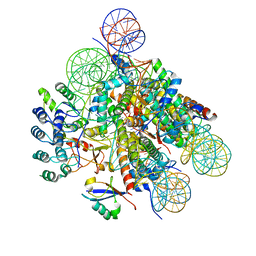 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | Authors: | Dai, Y, Dai, L, Zhou, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
6K08
 
 | | Crystal structure of REV7(R124A/A135D) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
6K07
 
 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKD
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UK5
 
 | |
7UKE
 
 | |
7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKG
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, transmembrane region | | Descriptor: | Dipeptidyl-peptidase 6, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKH
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, intracellular region | | Descriptor: | CALCIUM ION, Isoform 2 of Kv channel-interacting protein 2, Potassium voltage-gated channel subfamily D member 2, ... | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
8T69
 
 | | Human VMAT2 in complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T6B
 
 | | Human VMAT2 in complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T6A
 
 | | Human VMAT2 in complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8EX8
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX4
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX7
 
 | |
8EX5
 
 | |
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | Authors: | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
8JBI
 
 | | SteC 202-375 mutant- C276S | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Secreted effector kinase SteC | | Authors: | Zhang, M, Dai, Y, Li, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Salmonella manipulates macrophage cytoskeleton to penetrate gut-vascular barrier and promote dissemination during infection
To Be Published
|
|
8GPD
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxidanyl-2-oxidanylidene-1-(2-phenoxyethanoylamino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
