4FHR
 
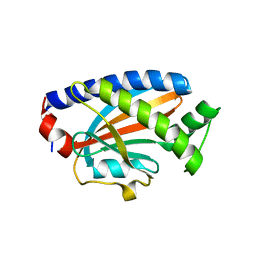 | | Crystal structure of the complex between the flagellar motor proteins FliG and FliM. | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Paz, A, Vartanian, A.S, Fortgang, E.A, Abramson, J, Dahlquist, F.W. | | Deposit date: | 2012-06-06 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Structure of flagellar motor proteins in complex allows for insights into motor structure and switching.
J.Biol.Chem., 287, 2012
|
|
2FSP
 
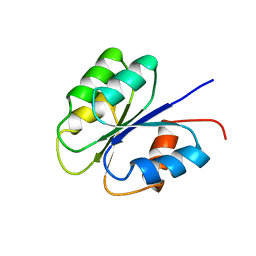 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
2TMY
 
 | |
3TMY
 
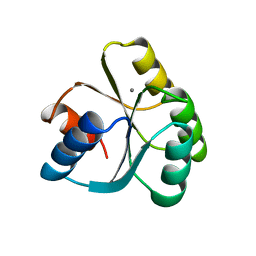 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
2HTJ
 
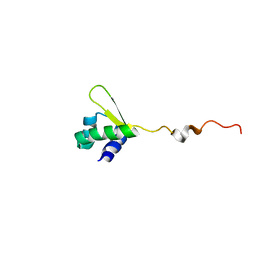 | | NMR structure of E.coli PapI | | Descriptor: | P fimbrial regulatory protein KS71A | | Authors: | Kawamura, T, Zhou, H, Le, L.U.K, Dahlquist, F.W. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-30 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Escherichia coli PapI, a Key Regulator of the Pap Pili Phase Variation.
J.Mol.Biol., 365, 2007
|
|
5TDY
 
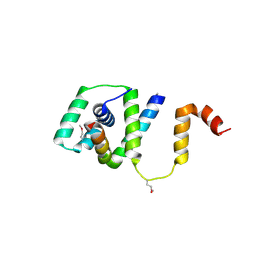 | | Structure of cofolded FliFc:FliGn complex from Thermotoga maritima | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG | | Authors: | Lynch, M.J, Levenson, R, Kim, E.A, Sircar, R, Blair, D.F, Dahlquist, F.W, Crane, B.R. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Structure, 25, 2017
|
|
4TMY
 
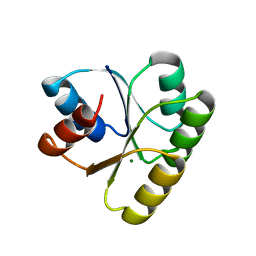 | | CHEY FROM THERMOTOGA MARITIMA (MG-IV) | | Descriptor: | CHEY PROTEIN, MAGNESIUM ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1T6O
 
 | | Nucleocapsid-binding domain of the measles virus P protein (amino acids 457-507) in complex with amino acids 486-505 of the measles virus N protein | | Descriptor: | linker, phosphoprotein | | Authors: | Kingston, R.L, Hamel, D.J, Gay, L.S, Dahlquist, F.W, Matthews, B.W. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the attachment of a paramyxoviral polymerase to its template.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U8T
 
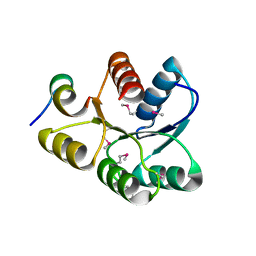 | | Crystal structure of CheY D13K Y106W alone and in complex with a FliM peptide | | Descriptor: | Chemotaxis protein cheY, Flagellar motor switch protein fliM, SULFATE ION | | Authors: | Dyer, C.M, Quillin, M.L, Campos, A, Lu, J, McEvoy, M.M, Hausrath, A.C, Westbrook, E.M, Matsumura, P, Matthews, B.W, Dahlquist, F.W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Constitutively Active Double Mutant CheY(D13K Y106W) Alone and in Complex with a FliM Peptide
J.Mol.Biol., 342, 2004
|
|
1TMY
 
 | |
2LD6
 
 | |
2LCB
 
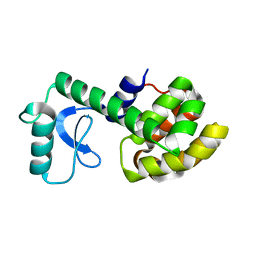 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-07 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-07 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2L4A
 
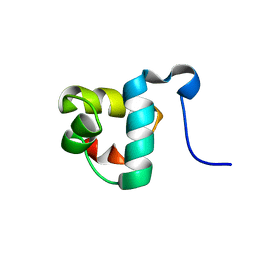 | |
2B1J
 
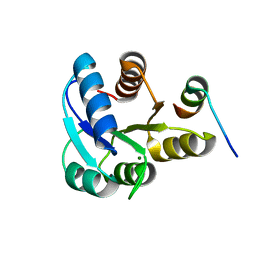 | |
1C4W
 
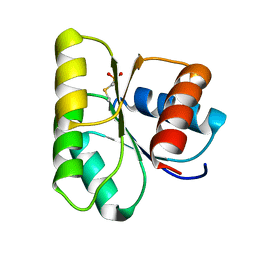 | | 1.9 A STRUCTURE OF A-THIOPHOSPHONATE MODIFIED CHEY D57C | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Halkides, C.J, McEvoy, M.M, Matsumura, P, Volz, K, Dahlquist, F.W. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY.
Biochemistry, 39, 2000
|
|
1FSP
 
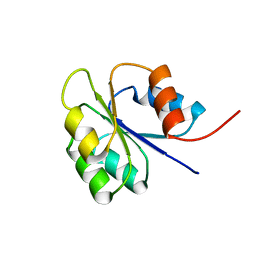 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, 20 STRUCTURES | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-05 | | Release date: | 1997-12-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1ABV
 
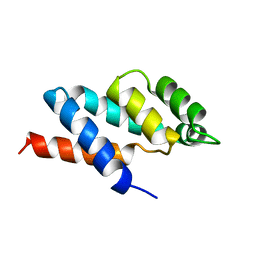 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | Authors: | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | Deposit date: | 1997-01-29 | | Release date: | 1997-07-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
1CEY
 
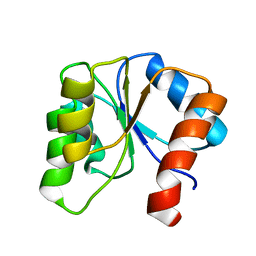 | | ASSIGNMENTS, SECONDARY STRUCTURE, GLOBAL FOLD, AND DYNAMICS OF CHEMOTAXIS Y PROTEIN USING THREE-AND FOUR-DIMENSIONAL HETERONUCLEAR (13C,15N) NMR SPECTROSCOPY | | Descriptor: | CHEY | | Authors: | Moy, F.J, Lowry, D.F, Matsumura, P, Dahlquist, F.W, Krywko, J.E, Domaille, P.J. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1DTM
 
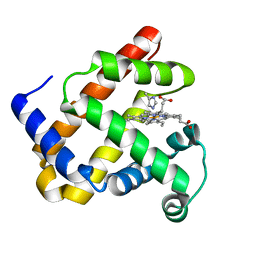 | |
1DUO
 
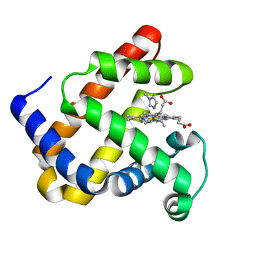 | |
1DUK
 
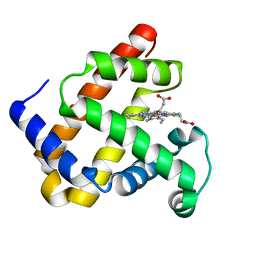 | | WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Authors: | Barrick, D, Dahlquist, F.W. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Trans-substitution of the proximal hydrogen bond in myoglobin: I. Structural consequences of hydrogen bond deletion.
Proteins, 39, 2000
|
|
1EAY
 
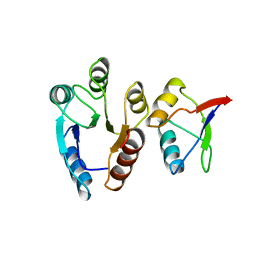 | | CHEY-BINDING (P2) DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEA, CHEY | | Authors: | Mcevoy, M.M, Hausrath, A.C, Randolph, G.B, Remington, S.J, Dahlquist, F.W. | | Deposit date: | 1998-04-23 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1F43
 
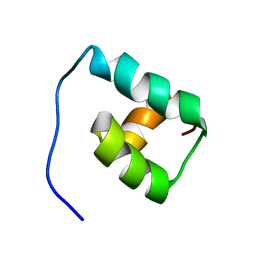 | | SOLUTION STRUCTURE OF THE MATA1 HOMEODOMAIN | | Descriptor: | MATING-TYPE PROTEIN A-1 | | Authors: | Anderson, J.S, Forman, M, Modleski, S, Dahlquist, F.W, Baxter, S.M. | | Deposit date: | 2000-06-07 | | Release date: | 2000-07-26 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Cooperative ordering in homeodomain-DNA recognition: solution structure and dynamics of the MATa1 homeodomain.
Biochemistry, 39, 2000
|
|
1FWP
 
 | |
