6OMI
 
 | |
6PIR
 
 | |
1MIR
 
 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1MFE
 
 | |
1AJ5
 
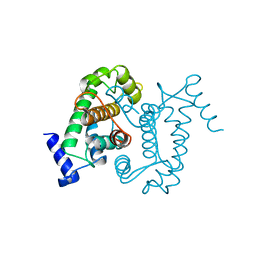 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
7KJ6
 
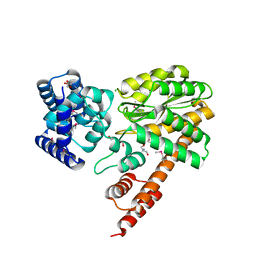 | | Structure of Legionella Effector LegA15 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ankyrin repeat-containing protein, CHLORIDE ION, ... | | Authors: | Cygler, M, Chung, I.Y.W. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-18 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Legionella effector LegA15/AnkH contains an unrecognized cysteine protease-like domain and displays structural similarity to LegA3/AnkD, but differs in host cell localization.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4Q5H
 
 | |
4Q5E
 
 | |
4II4
 
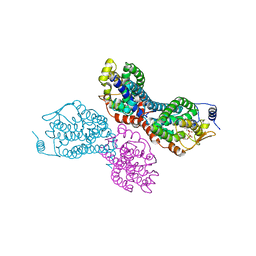 | |
4IIT
 
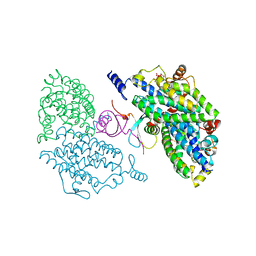 | |
6WO6
 
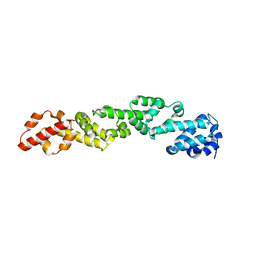 | |
6MCA
 
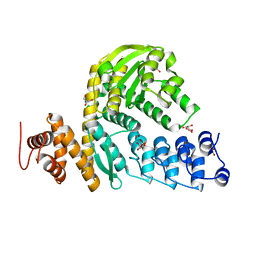 | | Structure of Legionella Effector AnkH | | Descriptor: | Ankyrin repeat domain protein, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Cygler, M, Chung, I.Y.W. | | Deposit date: | 2018-08-30 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Interaction of the Ankyrin H Core Effector ofLegionellawith the Host LARP7 Component of the 7SK snRNP Complex.
Mbio, 10, 2019
|
|
4LRJ
 
 | |
4LRK
 
 | |
1CS8
 
 | | CRYSTAL STRUCTURE OF PROCATHEPSIN L | | Descriptor: | HUMAN PROCATHEPSIN L | | Authors: | Cygler, M, Coulombe, R. | | Deposit date: | 1999-08-17 | | Release date: | 1999-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment.
EMBO J., 15, 1996
|
|
1DVI
 
 | | CALPAIN DOMAIN VI WITH CALCIUM BOUND | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Cygler, M, Blanchard, H, Grochulski, P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
1MFB
 
 | |
1MFC
 
 | |
3PVY
 
 | |
3PW1
 
 | |
3PVT
 
 | |
3PW8
 
 | |
3PWQ
 
 | |
3PVR
 
 | |
2FNI
 
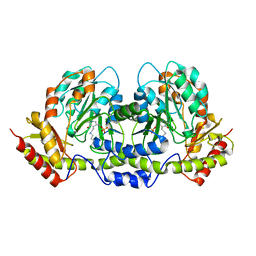 | | PseC aminotransferase involved in pseudoaminic acid biosynthesis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Cygler, M, Matte, A, Lunin, V.V, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
