1BJ5
 
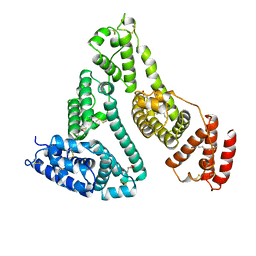 | | HUMAN SERUM ALBUMIN COMPLEXED WITH MYRISTIC ACID | | Descriptor: | HUMAN SERUM ALBUMIN, MYRISTIC ACID | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
1BKE
 
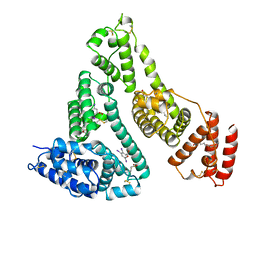 | | HUMAN SERUM ALBUMIN IN A COMPLEX WITH MYRISTIC ACID AND TRI-IODOBENZOIC ACID | | Descriptor: | 2,3,5-TRIIODOBENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
4GH4
 
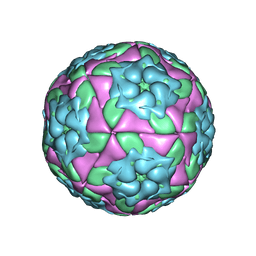 | | Crystal Structure of Foot and Mouth Disease Virus A22 Serotype | | Descriptor: | capsid protein VP1, capsid protein VP2, capsid protein VP3, ... | | Authors: | Kotecha, A, Jinshan, R, Curry, S, Fry, E, Stuart, D. | | Deposit date: | 2012-08-07 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Perturbations in the surface structure of A22 Iraq foot-and-mouth disease virus accompanying coupled changes in host cell specificity and antigenicity.
Structure, 4, 1996
|
|
4X2V
 
 | |
4X2W
 
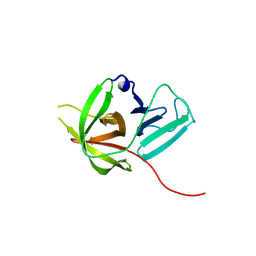 | |
4X2Y
 
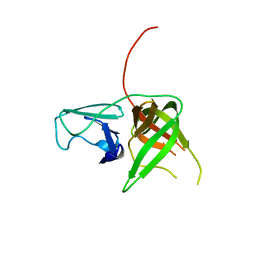 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
4X2X
 
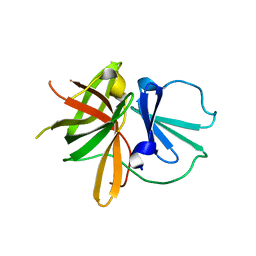 | |
5HM2
 
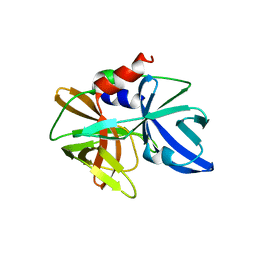 | |
2V6C
 
 | |
2J92
 
 | |
1AL2
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR7
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR8
 
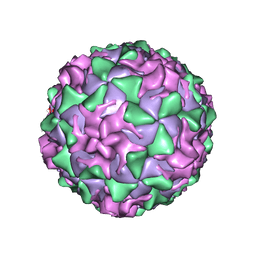 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
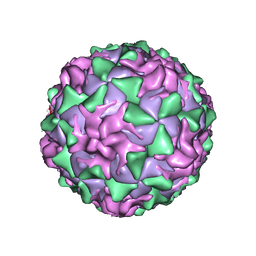 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR6
 
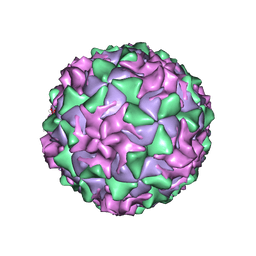 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR9
 
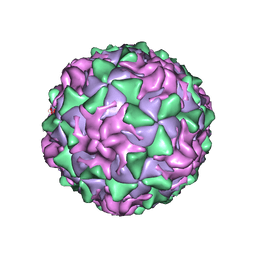 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1QM9
 
 | | NMR, REPRESENTATIVE STRUCTURE | | Descriptor: | POLYPYRIMIDINE TRACT-BINDING PROTEIN | | Authors: | Conte, M.R, Grune, T, Curry, S, Matthews, S. | | Deposit date: | 1999-09-22 | | Release date: | 2000-07-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of Tandem RNA Recognition Motifs from Polypyrimidine Tract Binding Protein Reveals Novel Features of the Rrm Fold
Embo J., 19, 2000
|
|
2J76
 
 | | Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4B | | Authors: | Fleming, K, Ghuman, J, Yuan, X.M, Simpson, P, Szendroi, A, Matthews, S, Curry, S. | | Deposit date: | 2006-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2017-04-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and RNA Interactions of the RNA Recognition Motif from Eukaryotic Translation Initiation Factor 4B.
Biochemistry, 42, 2003
|
|
2M4G
 
 | | Solution structure of the Core Domain (11-85) of the Murine Norovirus VPg protein | | Descriptor: | Murine Norovirus VPg protein | | Authors: | Leen, E.N, Kwok, R, Birtley, J.R, Prater, S.N, Simpson, P.J, Matthews, S, Marchant, J, Curry, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
2M4H
 
 | | Solution structure of the Core Domain (10-76) of the Feline Calicivirus VPg protein | | Descriptor: | Feline Calicivirus VPg protein | | Authors: | Kwok, R.N, Leen, E.N, Birtley, J.R, Prater, S.N, Simpson, P.J, Curry, S, Matthews, S, Marchant, J. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
1H9Z
 
 | |
1HA2
 
 | |
2VON
 
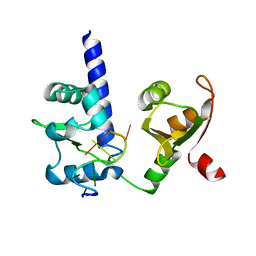 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUAAUUU | | Descriptor: | 5'-R(*AP*UP*AP*AP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
3ZZZ
 
 | |
3ZZY
 
 | |
