2HIL
 
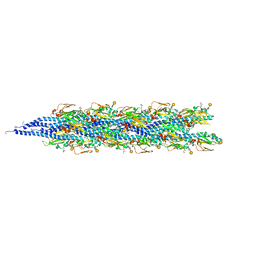 | | Structure of the Neisseria gonorrhoeae Type IV pilus filament from x-ray crystallography and electron cryomicroscopy | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4-dideoxy-beta-D-glucopyranose | | Authors: | Craig, L, Volkmann, N, Egelman, E.H, Tainer, J.A. | | Deposit date: | 2006-06-29 | | Release date: | 2006-09-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
2HI2
 
 | | Crystal structure of native Neisseria gonorrhoeae Type IV pilin at 2.3 Angstroms Resolution | | Descriptor: | Fimbrial protein, HEPTANE-1,2,3-TRIOL, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
1OQW
 
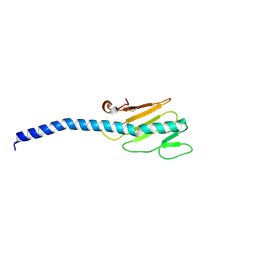 | |
1OQV
 
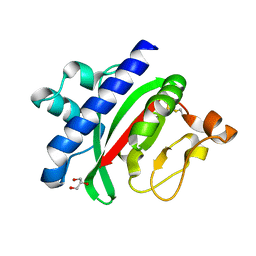 | |
3HRV
 
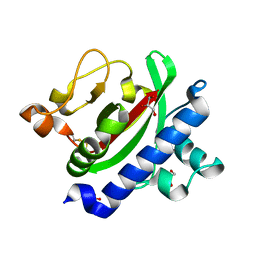 | | Crystal structure of TcpA, a Type IV pilin from Vibrio cholerae El Tor biotype | | Descriptor: | GLYCEROL, SULFATE ION, Toxin coregulated pilin | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation.
Mol.Microbiol., 77, 2010
|
|
3OC5
 
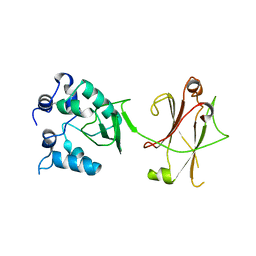 | |
3OC8
 
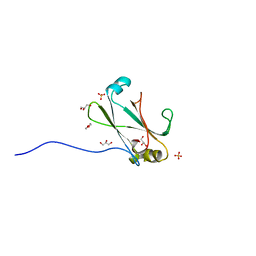 | | Crystal Structure of the C-terminal Domain of the Vibrio cholerae soluble colonization factor TcpF | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Craig, L, Kolappan, S, Yuen, A.S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Vibrio cholerae Colonization Factor TcpF and Identification of a Functional Immunogenic Site.
J.Mol.Biol., 409, 2011
|
|
4IJY
 
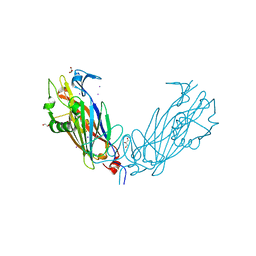 | | Crystal Structure of the ETEC Secreted Protein CofJ | | Descriptor: | CofJ, GLYCEROL, IODIDE ION | | Authors: | Craig, L, Kolappan, S, Yuen, A.S.W. | | Deposit date: | 2012-12-24 | | Release date: | 2013-10-16 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic Escherichia coli.
Mol.Microbiol., 90, 2013
|
|
4QSM
 
 | | Crystal structure of human muscle L-lactate dehydrogenase in complex with inhibitor 2, 3-{[7-(2,4-dimethoxypyrimidin-5-yl)-3-sulfamoylquinolin-4-yl]amino}benzoic acid | | Descriptor: | 3-{[7-(2,4-dimethoxypyrimidin-5-yl)-3-sulfamoylquinolin-4-yl]amino}benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-07-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5JW8
 
 | |
5KUA
 
 | | Cryo-EM reconstruction of Neisseria meningitidis Type IV pilus | | Descriptor: | pilin | | Authors: | Kolappan, S, Coureuil, M, Yu, X, Nassif, X, Craig, L, Egelman, E.H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-12 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Neisseria meningitidis Type IV pilus.
Nat Commun, 7, 2016
|
|
5EVG
 
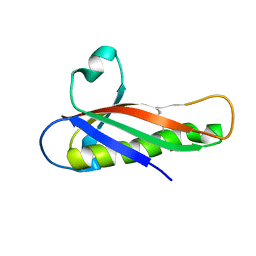 | | Crystal structure of a Francisella virulence factor FvfA in the orthorhombic form | | Descriptor: | Francisella virulence factor | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5EVF
 
 | | Crystal structure of a Francisella virulence factor FvfA in the hexagonal form | | Descriptor: | CHLORIDE ION, Francisella virulence factor, GLYCEROL | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1II8
 
 | | Crystal structure of the P. furiosus Rad50 ATPase domain | | Descriptor: | PHOSPHATE ION, Rad50 ABC-ATPase | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1II7
 
 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
8UHF
 
 | |
3S0T
 
 | |
5V0M
 
 | |
5V23
 
 | |
5VXX
 
 | | Cryo-EM reconstruction of Neisseria gonorrhoeae Type IV pilus | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose | | Authors: | Wang, F, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
5VXY
 
 | | Cryo-EM reconstruction of PAK pilus from Pseudomonas aeruginosa | | Descriptor: | Fimbrial protein | | Authors: | Wang, F, Osinksi, T, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
6MIC
 
 | |
4OJN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase A chain, PENTAETHYLENE GLYCOL | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OKN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase, ternary complex with NADH and oxalate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, KANAMYCIN A, L-lactate dehydrogenase A chain, ... | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QT0
 
 | | Crystal structure of human muscle L-lactate dehydrogenase in complex with inhibitor 1, 3-{[3-CARBAMOYL-7-(2,4-DIMETHOXYPYRIMIDIN-5-YL)QUINOLIN-4-YL]AMINO}BENZOIC ACID | | Descriptor: | 3-{[3-carbamoyl-7-(2,4-dimethoxypyrimidin-5-yl)quinolin-4-yl]amino}benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-07-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
