5PEP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEASES. II. THREE-DIMENSIONAL STRUCTURE OF THE HEXAGONAL CRYSTAL FORM OF PORCINE PEPSIN AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | PEPSIN | | Authors: | Cooper, J.B, Khan, G, Taylor, G, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-05-30 | | Release date: | 1990-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution.
J.Mol.Biol., 214, 1990
|
|
1IDS
 
 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
5ER1
 
 | |
2ER9
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES. | | Descriptor: | ENDOTHIAPEPSIN, L363,564 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
2ER0
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, L364,099 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
1GN6
 
 | | G152A mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Saward, S, Erskine, P.T, Badasso, M.O, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-05 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure Analysis of an Engineered Fe-Superoxide Dismutase Gly-Ala Mutant with Significantly Reduced Stability to Denaturant
FEBS Lett., 387, 1996
|
|
5OT1
 
 | | The type III pullulan hydrolase from Thermococcus kodakarensis | | Descriptor: | CALCIUM ION, Pullulanase type II, GH13 family | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Keegan, R, Ahmad, N, Muhammad, M.A, Rashid, N, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OV6
 
 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT0
 
 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OV4
 
 | | Bacillus megaterium porphobilinogen deaminase D82A mutant | | Descriptor: | Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1AW5
 
 | |
1B4E
 
 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
5L9A
 
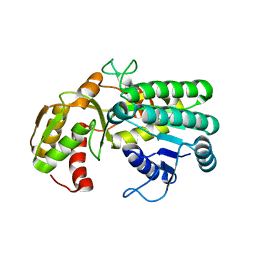 | | L-threonine dehydrogenase from trypanosoma brucei. | | Descriptor: | ACETATE ION, L-threonine 3-dehydrogenase | | Authors: | Erskine, P.T, Cooper, J.B, Adjogatse, E, Kelly, J, Wood, S.P. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LC1
 
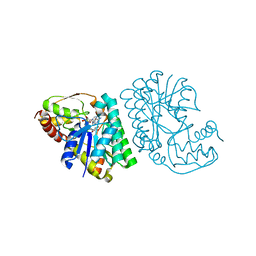 | | L-threonine dehydrogenase from Trypanosoma brucei with NAD and the inhibitor pyruvate bound. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, L-threonine 3-dehydrogenase, ... | | Authors: | Erskine, P.T, Adjogatse, E, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | Deposit date: | 2016-09-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MHB
 
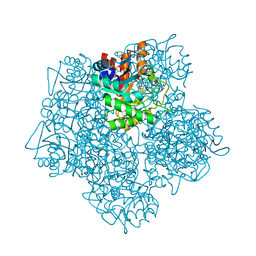 | | Product-Complex of E.coli 5-Amino Laevulinic Acid Dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, Delta-aminolevulinic acid dehydratase, GLYCEROL, ... | | Authors: | Norton, E, Erskine, P.T, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2016-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MDN
 
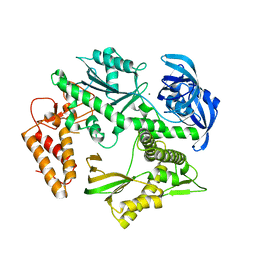 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OV5
 
 | | Bacillus megaterium porphobilinogen deaminase D82E mutant | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7P4R
 
 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
1EPO
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
1EPP
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPQ
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-133,450 (SOT PHE GLY+SCC GCL) | | Descriptor: | ENDOTHIAPEPSIN, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morpholin-4-ylsulfonyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Dealwis, C, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
5DZU
 
 | | Structure of potato cathepsin D inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease inhibitor 11, ... | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a Kunitz-type potato cathepsin D inhibitor.
J.Struct.Biol., 192, 2015
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5F6T
 
 | | Structure of calexcitin-Gd3+ complex. | | Descriptor: | CALCIUM ION, Calexcitin, GADOLINIUM ATOM | | Authors: | Chataigner, L, Guo, J, Erskine, P.T, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2015-12-06 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Binding of Gd(3+) to the neuronal signalling protein calexcitin identifies an exchangeable Ca(2+)-binding site.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
