1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1999-09-14 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
1VTY
 
 | | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs | | Descriptor: | AMINO GROUP, DNA (5'-D(*CP*(NH2)AP*CP*GP*TP*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs.
J. Biomol. Struct. Dyn., 4, 1986
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1H6F
 
 | | Human TBX3, a transcription factor responsible for ulnar-mammary syndrome, bound to a palindromic DNA site | | Descriptor: | 5'-D(*TP*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP* AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*T)-3', MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX3 | | Authors: | Coll, M, Muller, C.W. | | Deposit date: | 2001-06-13 | | Release date: | 2002-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx3, a Transcription Factor Responsible for Ulnar- Mammary Syndrome
Structure, 10, 2002
|
|
7P35
 
 | | Structure of the SARS-CoV-2 3CL protease in complex with rupintrivir | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Fabrega-Ferrer, M, Perez-Saavedra, J, Herrera-Morande, A, Coll, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
5NQ6
 
 | | Crystal structure of the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE from Escherichia coli at 2.40 Angstrom Resolution | | Descriptor: | GLYCEROL, SULFATE ION, Sulfur acceptor protein CsdE | | Authors: | Penya-Soler, E, Aranda, J, Lopez-Estepa, M, Gomez, S, Garces, F, Coll, M, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE.
PLoS ONE, 12, 2017
|
|
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
6EY7
 
 | | Human cytomegalovirus terminase nuclease domain, Mn soaked, inhibitor bound | | Descriptor: | 4-[(4-fluorophenyl)methyl-methyl-amino]-2,4-bis(oxidanylidene)butanoic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bongarzone, S, Nadal, M, Kaczmarska, Z, Machon, C, Alvarez, M, Albericio, F, Coll, M. | | Deposit date: | 2017-11-10 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Driven Discovery of alpha , gamma-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase.
Acs Omega, 3, 2018
|
|
1B01
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*GP*CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*GP*CP*AP*TP*TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*G)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
5N2Q
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to 26nt pMV158 oriT DNA | | Descriptor: | CHLORIDE ION, DNA (26-MER), GLYCEROL, ... | | Authors: | Russi, S, Boer, D.R, Coll, M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1D13
 
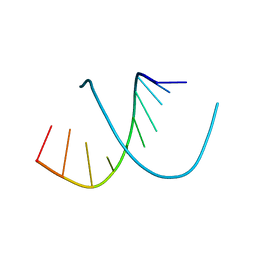 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
1CHN
 
 | |
1B00
 
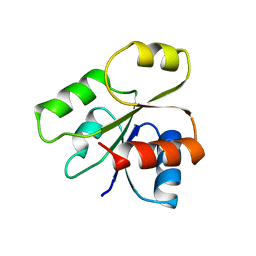 | | PHOB RECEIVER DOMAIN FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Sola, M, Gomis-Ruth, F.X, Serrano, L, Gonzalez, A, Coll, M. | | Deposit date: | 1998-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Three-dimensional crystal structure of the transcription factor PhoB receiver domain.
J.Mol.Biol., 285, 1999
|
|
1AYE
 
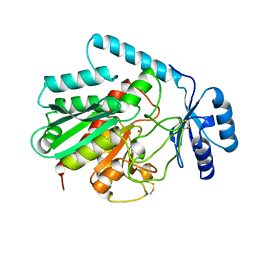 | | HUMAN PROCARBOXYPEPTIDASE A2 | | Descriptor: | PROCARBOXYPEPTIDASE A2, ZINC ION | | Authors: | Garcia-Saez, I, Reverte, D, Vendrell, J, Aviles, F.X, Coll, M. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of human procarboxypeptidase A2. Deciphering the basis of the inhibition, activation and intrinsic activity of the zymogen.
EMBO J., 16, 1997
|
|
2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
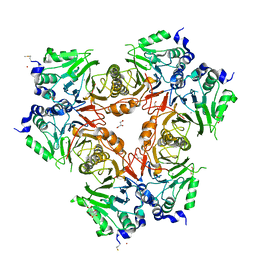 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
3I1D
 
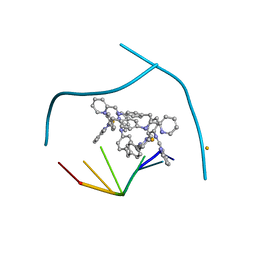 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | Deposit date: | 2009-06-26 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8B4D
 
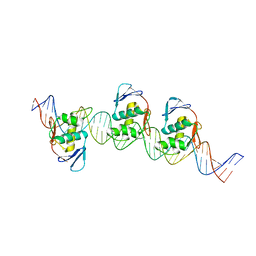 | | ToxR bacterial transcriptional regulator bound to 40 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (40-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4B
 
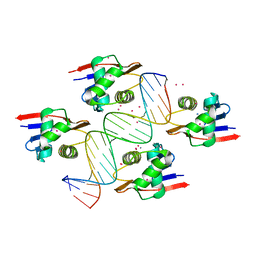 | | ToxR bacterial transcriptional regulator bound to 19 bp ompU promoter DNA | | Descriptor: | AMMONIUM ION, CADMIUM ION, Cholera toxin transcriptional activator, ... | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4C
 
 | | ToxR bacterial transcriptional regulator bound to 20 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (20-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4E
 
 | | ToxR bacterial transcriptional regulator bound to 25 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (25-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3FX8
 
 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2JB9
 
 | | PhoB response regulator receiver domain constitutively-active double mutant D10A and D53E. | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Ferrer-Orta, C, Arribas-Bosacoma, R, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the E. Coli Phob Receiver Domain Give Insights Into Activation
J.Mol.Biol., 366, 2007
|
|
