3G8C
 
 | | Crystal Structure of Biotin Carboxylase in Complex with Biotin, Bicarbonate, ADP and Mg Ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, BIOTIN, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
3G8D
 
 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
3RUP
 
 | |
3RV4
 
 | |
3RV3
 
 | |
5XU8
 
 | |
5XVE
 
 | |
5C3N
 
 | |
5Y3E
 
 | |
5Y3Q
 
 | |
2GPW
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, F363A Mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2GPS
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
5KS8
 
 | |
2OOE
 
 | | Crystal structure of HAT domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
2OND
 
 | | Crystal Structure of the HAT-C domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
7W1D
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a C2 crystal form | | Descriptor: | CARBONATE ION, CITRIC ACID, K1 LYASE | | Authors: | Tu, I.F, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
7W1E
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in complex with products | | Descriptor: | 2,6-anhydro-4,5-O-[(1R)-1-carboxyethylidene]-3-deoxy-L-threo-hex-2-enonic acid, 3-O-acetyl-6-deoxy-alpha-L-galactopyranose-(1-3)-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
7W1C
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a P1 crystal form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
4HI3
 
 | |
7F0U
 
 | |
5B7C
 
 | |
4RCN
 
 | |
4HNU
 
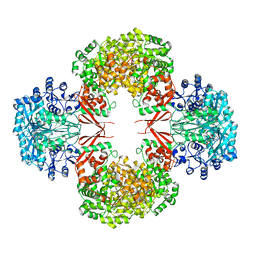 | | crystal structure of K442E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNV
 
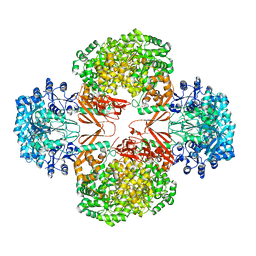 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNT
 
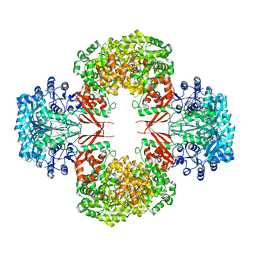 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
