2CHT
 
 | |
2CHS
 
 | |
1QBK
 
 | |
1COM
 
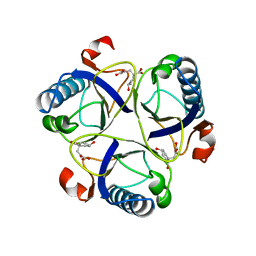 | | THE MONOFUNCTIONAL CHORISMATE MUTASE FROM BACILLUS SUBTILIS: STRUCTURE DETERMINATION OF CHORISMATE MUTASE AND ITS COMPLEXES WITH A TRANSITION STATE ANALOG AND PREPHENATE, AND IMPLICATIONS ON THE MECHANISM OF ENZYMATIC REACTION | | Descriptor: | CHORISMATE MUTASE, PREPHENIC ACID | | Authors: | Chook, Y.M, Ke, H, Lipscomb, W.N. | | Deposit date: | 1994-04-08 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction.
J.Mol.Biol., 240, 1994
|
|
2H4M
 
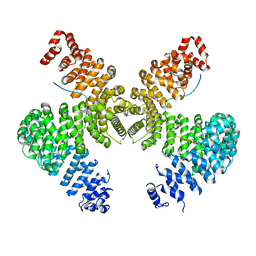 | | Karyopherin Beta2/Transportin-M9NLS | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, Transportin-1 | | Authors: | Chook, Y.M, Cansizoglu, A.E. | | Deposit date: | 2006-05-24 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rules for nuclear localization sequence recognition by karyopherin beta 2.
Cell(Cambridge,Mass.), 126, 2006
|
|
8DYO
 
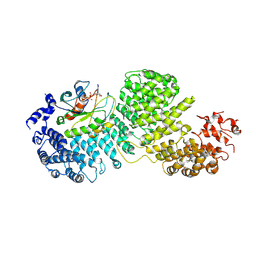 | | Cryo-EM structure of Importin-4 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | Authors: | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6N1Z
 
 | | Importin-9 bound to H2A-H2B | | Descriptor: | Histone H2A, Histone H2B 1.1, Importin-9 | | Authors: | Tomchick, D.R, Chook, Y.M, Padavannil, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone.
Elife, 8, 2019
|
|
3GB8
 
 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
7UNK
 
 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8SGH
 
 | |
7CYL
 
 | |
5J3V
 
 | |
5JLJ
 
 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
7L5E
 
 | |
4OO6
 
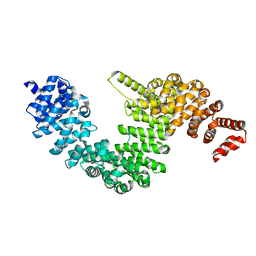 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
8F0X
 
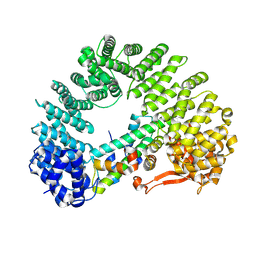 | | Cryo-EM structure of Kap114 bound to H2A-H2B | | Descriptor: | Histone H2A.2, Histone H2B.2, Importin subunit beta-5 | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F19
 
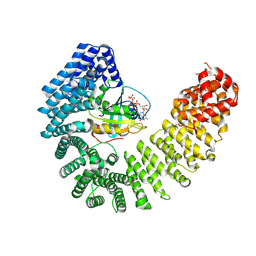 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-5, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F7A
 
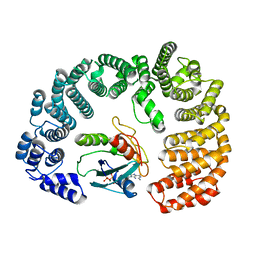 | | Cryo-EM structure of Importin-9 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-9, ... | | Authors: | Bernardes, N.E, Chook, Y.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1E
 
 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) and H2A-H2B | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Histone H2A.2, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2OT8
 
 | |
2QMR
 
 | | Karyopherin beta2/transportin | | Descriptor: | Transportin-1 | | Authors: | Cansizoglu, A.E, Chook, Y.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational heterogeneity of karyopherin beta2 is segmental
Structure, 15, 2007
|
|
6N34
 
 | | Crystal structure of the BTB domain of Human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B, Chook, Y.M. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N3H
 
 | | Crystal structure of Kelch domain of the human NS1 binding protein | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XJR
 
 | |
