2OS5
 
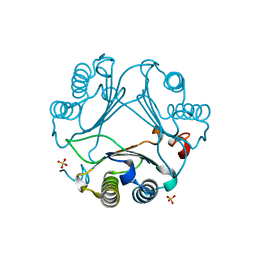 | | Macrophage migration inhibitory factor from Ancylostoma ceylanicum | | Descriptor: | AceMIF, SULFATE ION | | Authors: | Cho, Y, Lolis, E. | | Deposit date: | 2007-02-05 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of a secreted hookworm Macrophage Migration Inhibitory Factor (MIF) that interacts with the human MIF receptor CD74.
J.Biol.Chem., 282, 2007
|
|
1EFE
 
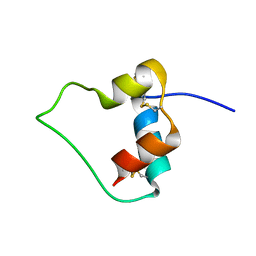 | | AN ACTIVE MINI-PROINSULIN, M2PI | | Descriptor: | MINI-PROINSULIN | | Authors: | Cho, Y, Chang, S.G, Choi, K.D, Shin, H, Ahn, B, Kim, K.S. | | Deposit date: | 2000-02-08 | | Release date: | 2000-03-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Active Mini-Proinsulin, M2PI: Inter-chain Flexibility is Crucial for Insulin Activity
J.Biochem.Mol.Biol., 33, 2000
|
|
1TSR
 
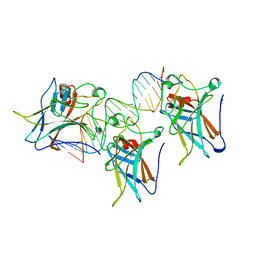 | | P53 CORE DOMAIN IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P, Pavletich, N. | | Deposit date: | 1995-07-28 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
1TUP
 
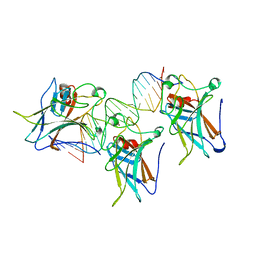 | | TUMOR SUPPRESSOR P53 COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR ), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1995-07-11 | | Release date: | 1995-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
3RF4
 
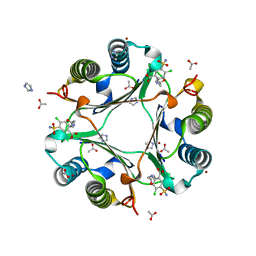 | | Ancylostoma ceylanicum mif in complex with furosemide | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Cho, Y, Lolis, E. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Drug Repositioning and Pharmacophore Identification in the Discovery of Hookworm MIF Inhibitors.
Chem.Biol., 18, 2011
|
|
3RF5
 
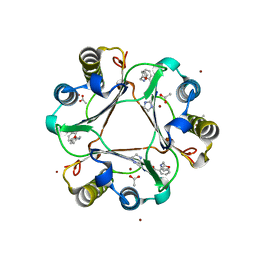 | | Ancylostoma ceylanicum mif in complex with n-(2,3,4,5,6-pentafluoro-benzyl)-4-sulfamoyl-benzamide | | Descriptor: | 2-[(furan-2-ylmethyl)amino]benzoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Cho, Y, Lolis, E. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug Repositioning and Pharmacophore Identification in the Discovery of Hookworm MIF Inhibitors.
Chem.Biol., 18, 2011
|
|
1NH7
 
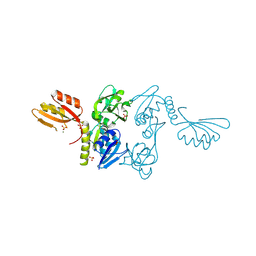 | | ATP PHOSPHORIBOSYLTRANSFERASE (ATP-PRTASE) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ATP Phosphoribosyltransferase, MAGNESIUM ION, SULFATE ION | | Authors: | Cho, Y, Sharma, V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ATP Phosphoribosyltransferase from Mycobacterium Tuberculosis
J.Biol.Chem., 278, 2003
|
|
1NH8
 
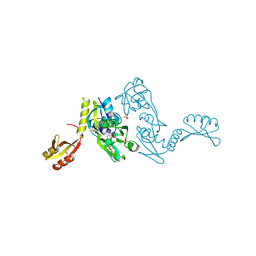 | | ATP PHOSPHORIBOSYLTRANSFERASE (ATP-PRTASE) FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AMP AND HISTIDINE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP Phosphoribosyltransferase, HISTIDINE, ... | | Authors: | Cho, Y, Sharma, V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ATP phosphoribosyltransferase from Mycobacterium tuberculosis
J.Biol.Chem., 278, 2003
|
|
2E6L
 
 | | structure of mouse WRN exonuclease domain | | Descriptor: | SULFATE ION, Werner syndrome ATP-dependent helicase homolog, ZINC ION | | Authors: | Cho, Y, Choi, J.M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | probing the roles of active site residues in 3'-5' exonuclease of werner syndrome protein
TO BE PUBLISHED
|
|
2E6M
 
 | | structure of mouse werner exonuclease domain | | Descriptor: | SULFATE ION, Werner syndrome ATP-dependent helicase homolog | | Authors: | Cho, Y, Choi, J.M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | probing the roles of active site residues in 3'-5' exonuclease of werner syndrome protein
TO BE PUBLISHED
|
|
4WFQ
 
 | | Crystal structure of TFIIH subunit | | Descriptor: | GLYCEROL, SULFATE ION, Suppressor of stem-loop protein 1 | | Authors: | Cho, Y, Kim, J.S, Lim, H.S. | | Deposit date: | 2014-09-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Rad3/XPD regulatory domain of Ssl1/p44
J.Biol.Chem., 290, 2015
|
|
7F4U
 
 | | Cryo-EM structure of TELO2-TTI1-TTI2 complex | | Descriptor: | TELO2-interacting protein 1 homolog, TELO2-interacting protein 2, Telomere length regulation protein TEL2 homolog | | Authors: | Cho, Y, Kim, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the Human TELO2-TTI1-TTI2 Complex.
J.Mol.Biol., 434, 2022
|
|
4R89
 
 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
4R8A
 
 | | Crystal structure of paFAN1 - 5' flap DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
3B6Q
 
 | |
3B6W
 
 | |
2ZXX
 
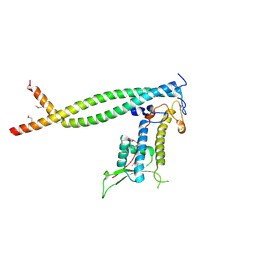 | | Crystal structure of Cdt1/geminin complex | | Descriptor: | DNA replication factor Cdt1, Geminin | | Authors: | Cho, Y, Lee, C, Hong, B.S, Choi, J.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the replication licensing factor Cdt1 by geminin
Nature, 430, 2004
|
|
2ZU6
 
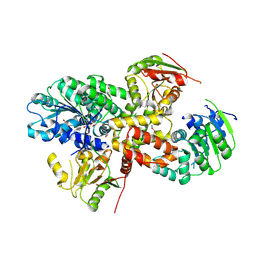 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3IJJ
 
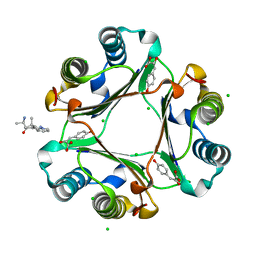 | | Ternary Complex of Macrophage Migration Inhibitory Factor (MIF) Bound Both to 4-hydroxyphenylpyruvate and to the Allosteric Inhibitor AV1013 (R-stereoisomer) | | Descriptor: | (2E)-2-hydroxy-3-(4-hydroxyphenyl)prop-2-enoic acid, (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ... | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IJG
 
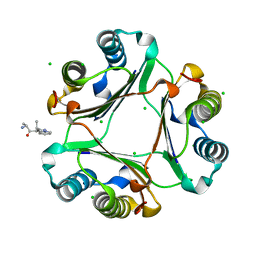 | | Macrophage Migration Inhibitory Factor (MIF) Bound to the (R)-Stereoisomer of AV1013 | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7DG2
 
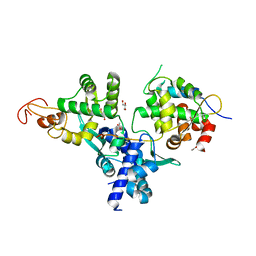 | | Nse1-Nse3-Nse4 complex | | Descriptor: | ACETATE ION, GLYCEROL, MAGE domain-containing protein, ... | | Authors: | Cho, Y, Jo, A. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure Basis for Shaping the Nse4 Protein by the Nse1 and Nse3 Dimer within the Smc5/6 Complex.
J.Mol.Biol., 433, 2021
|
|
5Y7G
 
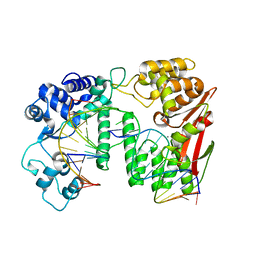 | | Crystal structure of paFAN1 bound to 1nt 5'flap DNA with gap | | Descriptor: | CALCIUM ION, DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Jin, H. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
2YT4
 
 | |
3A4C
 
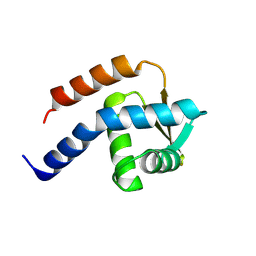 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
3B6T
 
 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Cho, Y, Lolis, E, Howe, J.R. | | Deposit date: | 2007-10-29 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
