1B8G
 
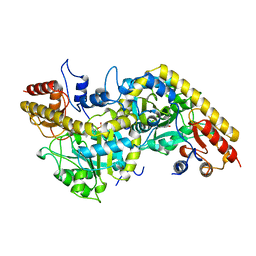 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
1F9M
 
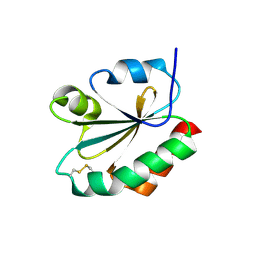 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (SHORT FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-11 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB0
 
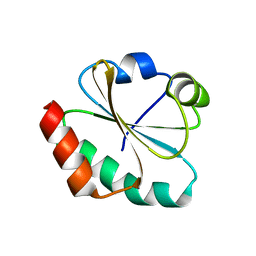 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (REDUCED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB6
 
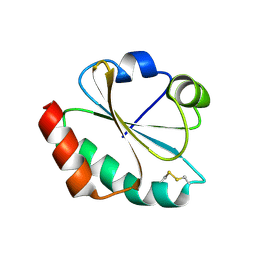 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (OXIDIZED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FAA
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (LONG FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-13 | | Release date: | 2000-09-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1YNU
 
 | | Crystal structure of apple ACC synthase in complex with L-vinylglycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ... | | Authors: | Capitani, G, Tschopp, M, Eliot, A.C, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of ACC synthase inactivated by the mechanism-based inhibitor L-vinylglycine.
Febs Lett., 579, 2005
|
|
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | Descriptor: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1JFU
 
 | | CRYSTAL STRUCTURE OF THE SOLUBLE DOMAIN OF TLPA FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN TLPA | | Authors: | Capitani, G, Rossmann, R, Sargent, D.F, Gruetter, M.G, Richmond, T.J, Hennecke, H. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties.
J.Mol.Biol., 311, 2001
|
|
1M4N
 
 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | Descriptor: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
1M7Y
 
 | | Crystal structure of apple ACC synthase in complex with L-aminoethoxyvinylglycine | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 1-aminocyclopropane-1-carboxylate synthase | | Authors: | Capitani, G, McCarthy, D, Gut, H, Gruetter, M.G, Kirsch, J.F. | | Deposit date: | 2002-07-23 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Apple 1-Aminocyclopropane-1-carboxylate Synthase in Complex with the Inhibitor
L-Aminoethoxyvinylglycine
J.Biol.Chem., 277, 2002
|
|
4WAS
 
 | | STRUCTURE OF THE ETR1P/NADP/CROTONYL-COA COMPLEX | | Descriptor: | CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Capitani, G, Erb, T.J. | | Deposit date: | 2014-08-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
3BFW
 
 | |
4DWH
 
 | | Structure of the major type 1 pilus subunit FIMA bound to the FIMC (2.5 A resolution) | | Descriptor: | Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Scharer, M.A, Puorger, C, Crespo, M, Glockshuber, R, Capitani, G. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
6SWH
 
 | | Crystal structure of the ternary complex between the type 1 pilus proteins FimC, FimI and FimA from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giese, C, Puorger, C, Ignatov, O, Weber, M, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7B0W
 
 | | Crystal structure of the E. coli type 1 pilus assembly inhibitor FimI bound to FimC | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, FORMIC ACID, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7B0X
 
 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | Authors: | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
3TQ7
 
 | | EB1c/EB3c heterodimer in complex with the CAP-Gly domain of P150glued | | Descriptor: | Dynactin subunit 1, Microtubule-associated protein RP/EB family member 1, Microtubule-associated protein RP/EB family member 3 | | Authors: | De Groot, C.O, Scharer, M.A, Capitani, G, Steinmetz, M.O. | | Deposit date: | 2011-09-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued).
J.Struct.Biol., 177, 2012
|
|
4TVV
 
 | | Crystal structure of LppA from Legionella pneumophila | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Weber, S, Stirnimann, C, Wieser, M, Meier, R, Engelhardt, S, Li, X, Capitani, G, Kammerer, R, Hilbi, H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Type IV Translocated Legionella Cysteine Phytase Counteracts Intracellular Growth Restriction by Phytate.
J.Biol.Chem., 289, 2014
|
|
4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2P2C
 
 | | Inhibition of caspase-2 by a designed ankyrin repeat protein (DARPin) | | Descriptor: | Caspase-2 | | Authors: | Roschitzki Voser, H, Briand, C, Capitani, G, Gruetter, M.G. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism.
Structure, 15, 2007
|
|
4P3M
 
 | | Crystal structure of serine hydroxymethyltransferase from Psychromonas ingrahamii | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Dworkowski, F, Angelaccio, S, Pascarella, S, Capitani, G. | | Deposit date: | 2014-03-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational transitions driven by pyridoxal-5'-phosphate uptake in the psychrophilic serine hydroxymethyltransferase from Psychromonas ingrahamii.
Proteins, 82, 2014
|
|
3MAF
 
 | | Crystal structure of StSPL (asymmetric form) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, sphingosine-1-phosphate lyase | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3PIU
 
 | |
