1AFX
 
 | |
1B36
 
 | |
1TLR
 
 | |
8TNS
 
 | |
5UQJ
 
 | | Structure of yeast Usb1 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Didychuk, A.L, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2017-02-08 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
5V1M
 
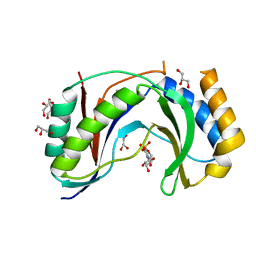 | | Structure of human Usb1 with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | Authors: | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
4N0T
 
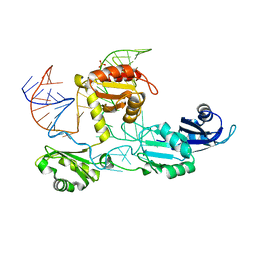 | | Core structure of the U6 small nuclear ribonucleoprotein at 1.7 Angstrom resolution | | Descriptor: | SULFATE ION, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA | | Authors: | Montemayor, E.J, Curran, E.C, Liao, H, Andrews, K.L, Treba, C.N, Butcher, S.E, Brow, D.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1Z2J
 
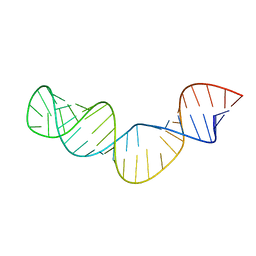 | |
2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
7MKT
 
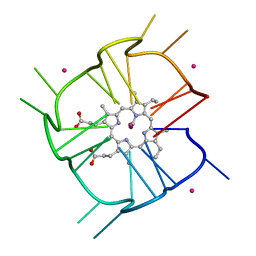 | | Crystal structure of r(GU)11G-NMM complex | | Descriptor: | N-METHYLMESOPORPHYRIN, POTASSIUM ION, RNA (5'-R(*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Bingman, C.A, Roschdi, S, Nomura, Y, Butcher, S.E. | | Deposit date: | 2021-04-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An atypical RNA quadruplex marks RNAs as vectors for gene silencing.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6ASO
 
 | | Structure of yeast U6 snRNP with 3'-phosphate terminated U6 RNA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Montemayor, E.J, Brow, D.A, Butcher, S.E. | | Deposit date: | 2017-08-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Architecture of the U6 snRNP reveals specific recognition of 3'-end processed U6 snRNA.
Nat Commun, 9, 2018
|
|
6D31
 
 | | Structure of human Usb1 with adenosine 5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nomura, Y, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-04-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and mechanistic basis for preferential deadenylation of U6 snRNA by Usb1.
Nucleic Acids Res., 46, 2018
|
|
5TF6
 
 | | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Montemayor, E.J, Brow, D.A, Butcher, S.E. | | Deposit date: | 2016-09-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6E7L
 
 | | Structure of a dimerized UUCG motif | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*UP*GP*CP*CP*C)-3') | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-07-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of an RNA helix with pyrimidine mismatches and cross-strand stacking.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1SY4
 
 | | Refined solution structure of the S. cerevisiae U6 INTRAMOLECULAR STEM LOOP (ISL) RNA USING RESIDUAL DIPOLAR COUPLINGS (RDCS) | | Descriptor: | U6 Intramolecular Stem-Loop RNA | | Authors: | Reiter, N.J, Nikstad, L.J, Allman, A.M, Johnson, R.J, Butcher, S.E. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the U6 RNA intramolecular stem-loop harboring an
S(P)-phosphorothioate modification.
RNA, 9, 2003
|
|
1SYZ
 
 | |
2KEZ
 
 | | NMR structure of U6 ISL at pH 8.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
2KF0
 
 | | NMR structure of U6 ISL at pH 7.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
6PPN
 
 | | Structure of S. pombe Lsm2-8 with unprocessed U6 snRNA | | Descriptor: | Mimic of unprocessed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
6PFQ
 
 | |
6PGL
 
 | | Structure of Kluyveromyces marxianus Usb1 with uridine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, GLYCEROL, ... | | Authors: | Nomura, Y, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis for the evolution of cyclic phosphodiesterase activity in the U6 snRNA exoribonuclease Usb1.
Nucleic Acids Res., 48, 2020
|
|
6PPQ
 
 | |
6PPV
 
 | |
6PPP
 
 | | Structure of S. pombe Lsm2-8 with processed U6 snRNA | | Descriptor: | Mimic of processed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
2N7M
 
 | | NMR-SAXS/WAXS Structure of the core of the U4/U6 di-snRNA | | Descriptor: | RNA (92-MER) | | Authors: | Cornilescu, G, Didychuk, A.L, Rodgers, M.L, Michael, L.A, Burke, J.E, Montemayor, E.J, Hoskins, A.A, Butcher, S.E, Tonelli, M. | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA.
J.Mol.Biol., 428, 2016
|
|
