233D
 
 | | THE CRYSTAL STRUCTURE ANALYSIS OF D(CGCGAASSCGCG)2: A SYNTHETIC DNA DODECAMER DUPLEX CONTAINING FOUR 4'-THIO-2'-DEOXYTHYMIDINE NUCLEOTIDES | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*)-D(*(T49)P*(T49)P*)-D(*CP*GP*CP*G)-3') | | Authors: | Boggon, T.J, Hancox, E.L, McAuley-Hecht, K.E, Connolly, B.A, Hunter, W.N, Brown, T, Walker, R.T, Leonard, G.A. | | Deposit date: | 1995-10-16 | | Release date: | 1996-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure analysis of d(CGCGAASSCGCG)2, a synthetic DNA dodecamer duplex containing four 4'-thio-2'-deoxythymidine nucleotides.
Nucleic Acids Res., 24, 1996
|
|
1YVJ
 
 | | Crystal structure of the Jak3 kinase domain in complex with a staurosporine analogue | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Tyrosine-protein kinase JAK3 | | Authors: | Boggon, T.J, Li, Y, Manley, P.W, Eck, M.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog
Blood, 106, 2005
|
|
1L3W
 
 | | C-cadherin Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Boggon, T.J, Murray, J, Chappuis-Flament, S, Wong, E, Gumbiner, B.M, Shapiro, L. | | Deposit date: | 2002-03-01 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | C-cadherin ectodomain structure and implications for cell adhesion mechanisms
Science, 296, 2002
|
|
1C8Z
 
 | |
8DGQ
 
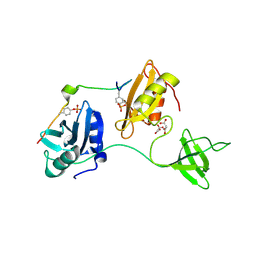 | |
4WJ7
 
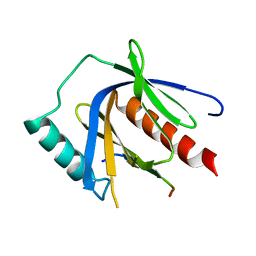 | | CCM2 PTB domain in complex with KRIT1 NPxY/F3 | | Descriptor: | KRIT1 NPxY/F3, Malcavernin | | Authors: | Fisher, O.S, Liu, W, Zhang, R, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structural Basis for the Disruption of the Cerebral Cavernous Malformations 2 (CCM2) Interaction with Krev Interaction Trapped 1 (KRIT1) by Disease-associated Mutations.
J.Biol.Chem., 290, 2015
|
|
8GI4
 
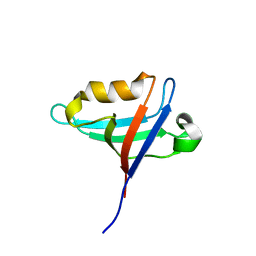 | |
3F6Q
 
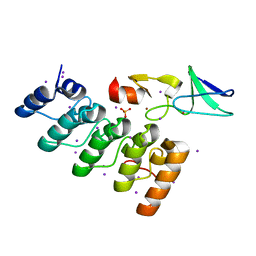 | | Crystal structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain | | Descriptor: | IODIDE ION, Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Chiswell, B.P, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of integrin-linked kinase-PINCH interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6PXB
 
 | |
4TKN
 
 | | Structure of the SNX17 FERM domain bound to the second NPxF motif of KRIT1 | | Descriptor: | Krev interaction trapped protein 1, Sorting nexin-17 | | Authors: | Stiegler, A.L, Zhang, R, Liu, W, Boggon, T.J. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 289, 2014
|
|
4TVQ
 
 | | CCM3 in complex with CCM2 LD-like motif | | Descriptor: | Cerebral cavernous malformations 2 protein, Cerebral cavernous malformations 3 protein | | Authors: | Li, X, Zhang, R, Fisher, O.S, Boggon, T.J. | | Deposit date: | 2014-06-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation.
J.Cell Biol., 208, 2015
|
|
6PXC
 
 | |
5HVJ
 
 | |
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
5HVK
 
 | |
4PGZ
 
 | | Structural basis of KIT activation by oncogenic mutations in the extracellular region reveals a zipper-like mechanism for ligand-dependent or oncogenic receptor tyrosine kinase activation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Mast/stem cell growth factor receptor Kit | | Authors: | Reshetnyak, A.V, Boggon, T.J, Lax, I, Schlessinger, J. | | Deposit date: | 2014-05-03 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer.
Mol.Cell, 57, 2015
|
|
7LNY
 
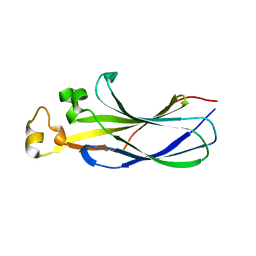 | |
7LO0
 
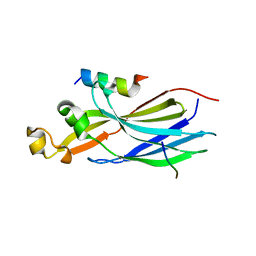 | | Structure of human ASF1a in complex with a TLK2 peptide | | Descriptor: | Histone chaperone ASF1A, Serine/threonine-protein kinase tousled-like 2 | | Authors: | Simon, B, Calderwood, D, Turk, B.E, Boggon, T.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Tousled-like kinase 2 targets ASF1 histone chaperones through client mimicry.
Nat Commun, 13, 2022
|
|
6BHC
 
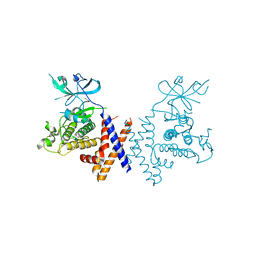 | | Crystal structure of pseduokinase PEAK1 (Sugen Kinase 269) | | Descriptor: | Pseudopodium-enriched atypical kinase 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of pseudokinase PEAK1 (Sugen kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization.
J. Biol. Chem., 293, 2018
|
|
6D4G
 
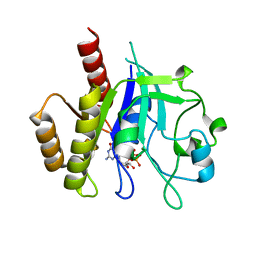 | | N-GTPase domain of p190RhoGAP-A | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 35 | | Authors: | Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2018-04-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The N-Terminal GTPase Domain of p190RhoGAP Proteins Is a PseudoGTPase.
Structure, 26, 2018
|
|
3TH5
 
 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3ULR
 
 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | Descriptor: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | Authors: | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2011-11-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1OBU
 
 | | Apocrustacyanin C1 crystals grown in space and earth using vapour diffusion geometry | | Descriptor: | CRUSTACYANIN C1 SUBUNIT | | Authors: | Habash, J, Boggon, T.J, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apocrustacyanin C(1) Crystals Grown in Space and on Earth Using Vapour-Diffusion Geometry: Protein Structure Refinements and Electron-Density Map Comparisons
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OBQ
 
 | | Apocrustacyanin C1 crystals grown in space and earth using vapour diffusion geometry | | Descriptor: | CRUSTACYANIN C1 SUBUNIT | | Authors: | Habash, J, Boggon, T.J, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Apocrustacyanin C(1) Crystals Grown in Space and on Earth Using Vapour-Diffusion Geometry: Protein Structure Refinements and Electron-Density Map Comparisons
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7TPB
 
 | | p120RasGAP SH3 domain in complex with DLC1 RhoGAP domain | | Descriptor: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 7 | | Authors: | Stiegler, A.L, Boggon, T.J, Chau, J.E, Vish, K.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SH3 domain regulation of RhoGAP activity: Crosstalk between p120RasGAP and DLC1 RhoGAP.
Nat Commun, 13, 2022
|
|
