5DTR
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD5 [N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTQ
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [(2,6-dichlorophenyl)(quinolin-6-yl)methanone] | | Descriptor: | (2,6-dichlorophenyl)(quinolin-6-yl)methanone, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTM
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide] | | Descriptor: | 4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
6UD7
 
 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | Authors: | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | Descriptor: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
5NTP
 
 | |
5NTQ
 
 | |
5NTI
 
 | |
5NTK
 
 | | Structural states of RORgt: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds | | Descriptor: | Nuclear receptor ROR-gamma, [(3~{S},8~{S},9~{S},10~{R},13~{R},14~{S},17~{R})-10,13-dimethyl-17-[(2~{R})-5-[[4-(2-morpholin-4-ylethoxy)phenyl]methylamino]-5-oxidanylidene-pentan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] hydrogen sulfate | | Authors: | Kallen, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural States of ROR gamma t: X-ray Elucidation of Molecular Mechanisms and Binding Interactions for Natural and Synthetic Compounds.
ChemMedChem, 12, 2017
|
|
5NTN
 
 | | Structural states of RORgt: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds | | Descriptor: | (5R,10S,13R,14R,17R)-17-((R,E)-7-hydroxy-6-methylhept-5-en-2-yl)-4,4,10,13,14-pentamethyl-1,2,5,6,10,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthrene-3,7(4H)-dione, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural States of ROR gamma t: X-ray Elucidation of Molecular Mechanisms and Binding Interactions for Natural and Synthetic Compounds.
ChemMedChem, 12, 2017
|
|
5NTW
 
 | |
6TCZ
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 | | Descriptor: | Proteasome endopeptidase complex, Proteasome subunit alpha type, Proteasome subunit beta, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
6TD5
 
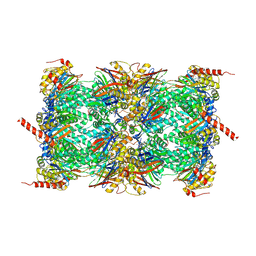 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
5NU1
 
 | |
3M42
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a tetracyclic ATP site inhibitor | | Descriptor: | 2-[5-(2-methoxyethoxy)pyridin-3-yl]-8,9,10,11-tetrahydro-7H-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin-7-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Scheufler, C, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Kroemer, M. | | Deposit date: | 2010-03-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel pyrrolo[2,3-f]isoquinoline based MAPKAP-K2 (MK2) inhibitors with potent in vitro and in vivo activity
To be Published
|
|
8AYH
 
 | |
5DRT
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD2 [2-(2-(5-((2-chlorophenoxy)methyl)-1H-tetrazol-1-yl)acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide] | | Descriptor: | 2-({5-[(2-chlorophenoxy)methyl]-1H-tetrazol-1-yl}acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DSX
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD10 [6'-chloro-1,4-dimethyl-5'-(2-methyl-6-((4-(methylamino)pyrimidin-2-yl)amino)-1H-indol-1-yl)-[3,3'-bipyridin]-2(1H)-one] | | Descriptor: | 6'-chloro-1,4-dimethyl-5'-(2-methyl-6-{[4-(methylamino)pyrimidin-2-yl]amino}-1H-indol-1-yl)-3,3'-bipyridin-2(1H)-one, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DT2
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DRY
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
6SJ7
 
 | |
6GG0
 
 | |
