1A5R
 
 | | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | | Descriptor: | SUMO-1 | | Authors: | Bayer, P, Arndt, A, Metzger, S, Mahajan, R, Melchior, F, Jaenicke, R, Becker, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the small ubiquitin-related modifier SUMO-1.
J.Mol.Biol., 280, 1998
|
|
1TIV
 
 | |
1BWX
 
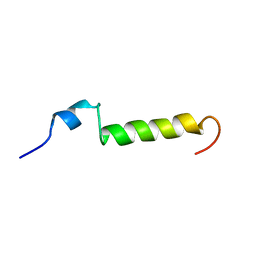 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-39, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-29 | | Release date: | 2000-01-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
1NMW
 
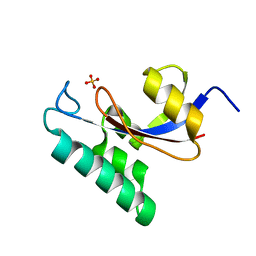 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NMV
 
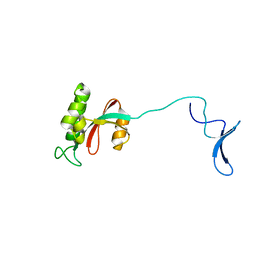 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1EQ3
 
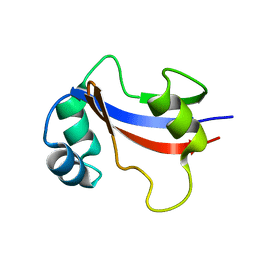 | | NMR STRUCTURE OF HUMAN PARVULIN HPAR14 | | Descriptor: | PEPTIDYL-PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Sekerina, E, Rahfeld, U.J, Muller, J, Fischer, G, Bayer, P. | | Deposit date: | 2000-04-02 | | Release date: | 2001-04-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of hPar14 reveals similarity to the peptidyl prolyl cis/trans isomerase domain of the mitotic regulator hPin1 but indicates a different functionality of the protein.
J.Mol.Biol., 301, 2000
|
|
1HPY
 
 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-34 IN 20% TRIFLUORETHANOL, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-30 | | Release date: | 2000-01-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
6GMP
 
 | | CRYSTAL STRUCTURE OF THE PPIASE DOMAIN OF TBPAR42 | | Descriptor: | PARVULIN 42 | | Authors: | Hoenig, D, Rute, A, Hofmann, E, Bayer, P, Gasper, R. | | Deposit date: | 2018-05-28 | | Release date: | 2019-03-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of the 42 kDa Parvulin ofTrypanosoma brucei.
Biomolecules, 9, 2019
|
|
1N3G
 
 | |
3UI4
 
 | | 0.8 A resolution crystal structure of human Parvulin 14 | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SULFATE ION | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
3UI6
 
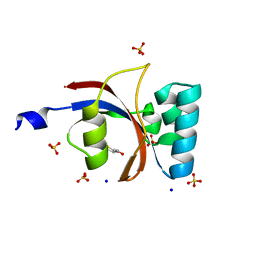 | | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT
To be Published
|
|
3UI5
 
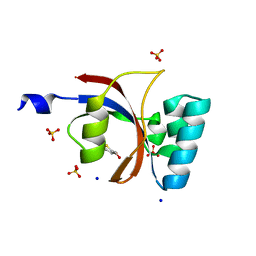 | | Crystal structure of human Parvulin 14 | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
2FNF
 
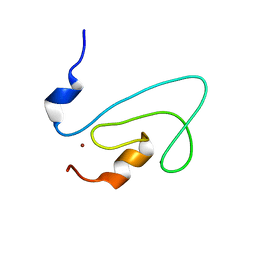 | | C1 domain of Nore1 | | Descriptor: | ZINC ION, putative Ras Effector Nore1 | | Authors: | Harjes, E, Harjes, S, Wohlgemuth, S, Krieger, E, Herrmann, C, Muller, K.H, Bayer, P. | | Deposit date: | 2006-01-11 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1.
Structure, 14, 2006
|
|
6SAP
 
 | |
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
2NAR
 
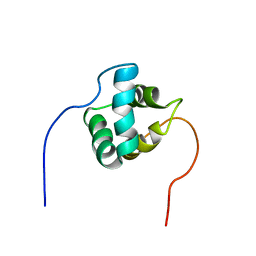 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2018-06-06 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
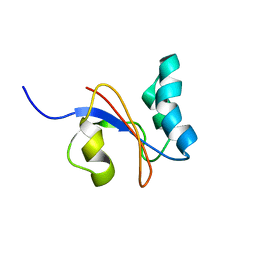
2M08
 
 | |
1U1W
 
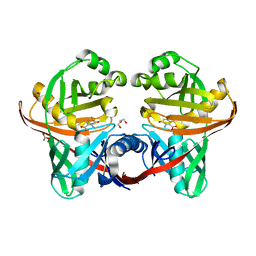 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1X
 
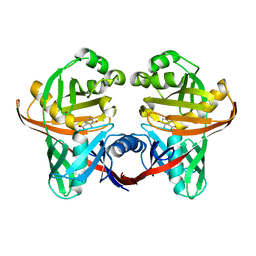 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1V
 
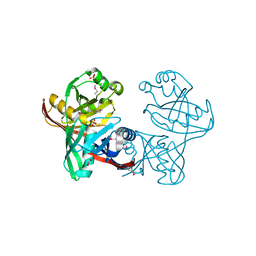 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XUB
 
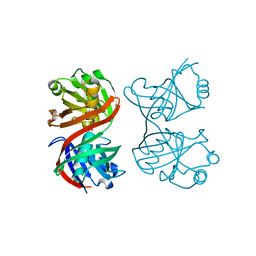 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4AY8
 
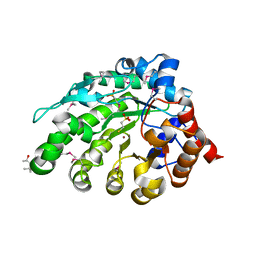 | | SeMet-derivative of a methyltransferase from M. mazei | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-THIOETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeldt, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AY7
 
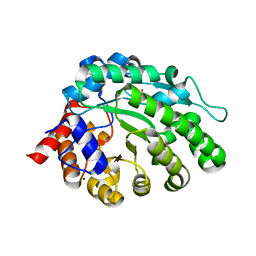 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1RFH
 
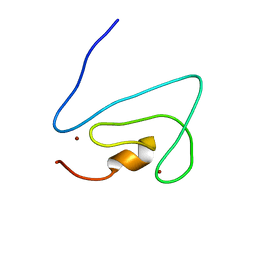 | | Solution structure of the C1 domain of Nore1, a novel Ras effector | | Descriptor: | Ras association (RalGDS/AF-6) domain family 5, ZINC ION | | Authors: | Guiberman, E, Wohlgemuth, S, Herrmann, C, Harjes, S, Mueller, K.H, Bayer, P. | | Deposit date: | 2003-11-09 | | Release date: | 2005-06-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of C1 domain of Nore1
To be Published
|
|
